Figure 1. Nitr13 and Nitr14 sequences.
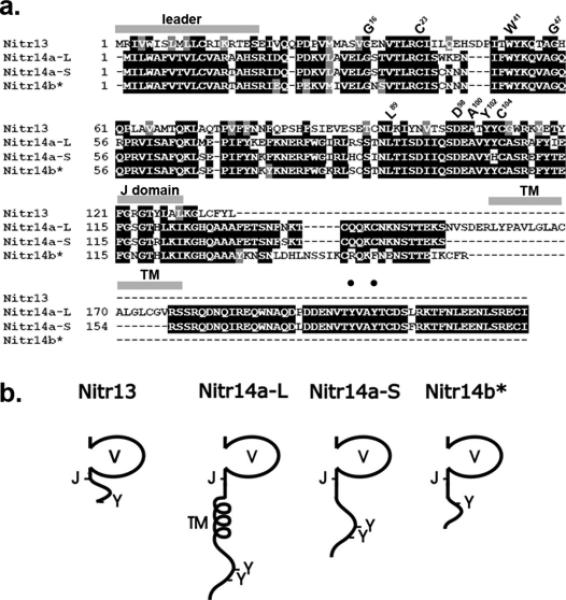
(a) The peptide sequences encoded by the nitr13 (Nitr13), nitr14a-Long (Nitr14a-L) and Nitr14a-Short (Nitr14a-S) cDNAs are aligned with the Nitr14b peptide sequence predicted from genomic sequence. Black shading indicates identical residues whereas gray shading indicates functionally similar residues. The locations of the predicted leader peptide sequences, transmembrane domains (TM), and joining (J) domains (FGXGTXLX(L/I)) are indicated with gray bars above the alignment. Residues, which are highly conserved in Ig domains (Litman et al. 2001), are indicated above the alignment using the IMGT numbering system (Giudicelli et al. 2006). Black circles indicate the locations of two tyrosines present in the cytoplasmic tail of Nitr14aL. (b) Predicted protein structures for Nitr13 and members of the Nitr14 family. Variable (V), transmembrane (TM), joining (J) and carboxy-terminal tyrosines (Y) are indicated. Only Nitr14a-L includes a transmembrane domain. The asterisk (*) next to Nitr14b indicates that this sequence is predicted from genomic sequence.
