Fig. 1.
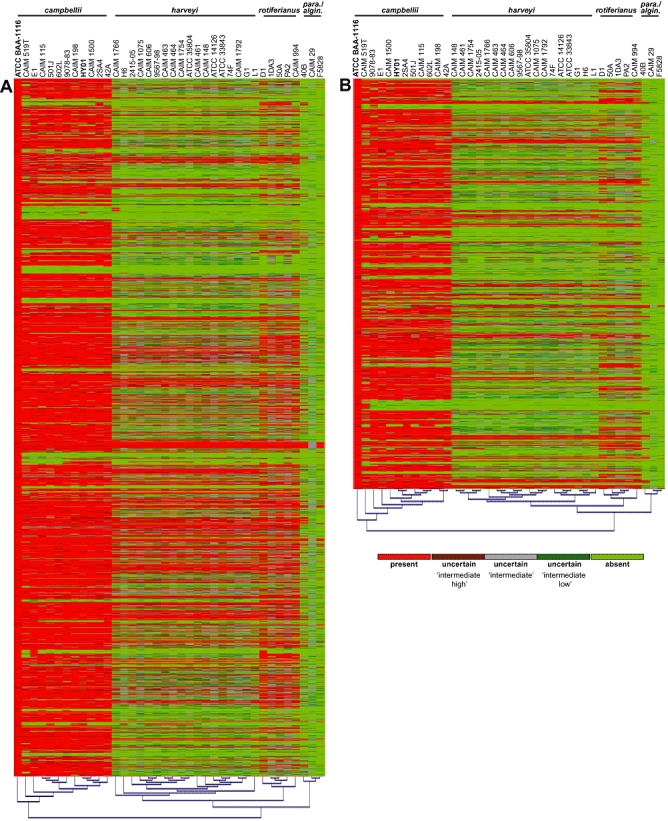
Hierarchically clustered heat maps based on CGH profiles demonstrating the presence and absence of genes within Harveyi clade members with respect to V. harveyi BAA-1116. A. Chromosome I, 2999 CDS. B. Chromosome II, 1765 CDS. The CDS in each heat map are ordered according to the genome structure of strain BAA-1116. Each CDS is depicted by one of five possible hybridization states (scale bar): (i) positive hybridization (CDS present call) = bright red bars, (ii) between positive and intermediate hybridization (uncertain ‘intermediate high’ call) = dark red bars, (iii) intermediate hybridization (uncertain ‘intermediate’ call) = grey bars, (iv) between intermediate and no hybridization (uncertain ‘intermediate low’ call) = dark green bars, and (v) no hybridization (CDS absent call) = bright green bars. Presence/absence designations generated from the hybridization profiles were calculated using the avgdiff method and clustered and visualized using MultiExperiment Viewer (MeV v4.4) software. ‘Para./algin.’ = parahaemolyticus and alginolyticus subclade.
