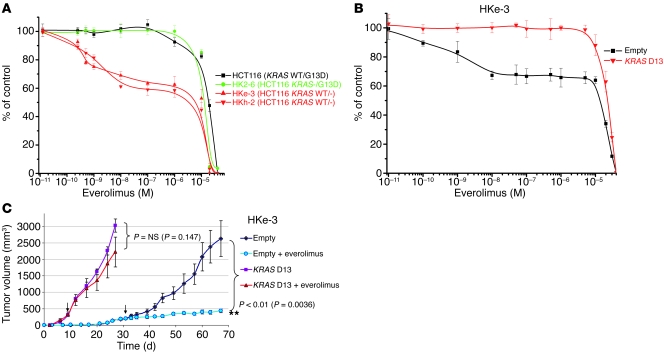
Figure 2. Oncogenic KRAS D13 confers resistance to everolimus.
(A) Two independent clones of HCT116 colorectal cancer cells — in which the KRAS D13 allele was genetically deleted by homologous recombination (HKh-2 and HKe-3, depicted in red) — were more sensitive to everolimus than either their parental cells (black) or a clone in which the KRAS WT allele was knocked out but the mutated allele was retained (HK2-6, green). Data are mean ± SD of at least 3 independent observations. (B) Effect of everolimus (96 hours) on proliferation of HKe-3 (HCT116-derivative KRAS WT clone) cells infected with control or KRAS D13 lentivirus. Results are expressed as percent viability compared with cells treated with DMSO only (control) and represent mean ± SD of at least 3 independent observations. (C) NOD/SCID mice were inoculated with HKe-3 cells (5 × 106) transduced with empty or KRAS D13 lentiviral vectors; once tumors were established, animals were administered everolimus at 7.5 mg/kg every other day. The arrows indicate the time point at which drug treatment was started. Results are shown as mean ± SEM.