Figure 6. Minimax contig sizes observed for simulated viral metagenome assemblies.
For a viral metagenome experiment design based on a Poisson number of species, uniformly distributed genome sizes and Pareto distributed abundances (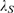 = 100,
= 100,  = Uniform(50000,350000),
= Uniform(50000,350000), 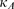 = 3.5),
= 3.5), 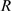 = 67109, 96992 and 126271 were calculated to have 95% probability of yielding assembled contigs of at least size
= 67109, 96992 and 126271 were calculated to have 95% probability of yielding assembled contigs of at least size  = 4, 5 and 6 for all species respectively. In Fig. 6, we show the distribution of minimax contig sizes obtained from 100 simulations of an assembly of these numbers of reads on a pool of
= 4, 5 and 6 for all species respectively. In Fig. 6, we show the distribution of minimax contig sizes obtained from 100 simulations of an assembly of these numbers of reads on a pool of  = 100 species with Uniform(50000,350000)-distributed genome sizes and Pareto(1,3.5)-distributed abundances (solid lines) vs. their targeted sizes (dashed). Consistent with previous observations for this case, the actual contig sizes obtained are slightly smaller than the targeted length. The median minimax contig sizes are 3.68, 4.85 and 6.16 (in read lengths, which is 92–103% of the target length), and 95% of all experiments yield contigs of length 3.38, 4.43 and 5.63 from all species (85–94% of the target length). The slight undersizing of contigs is consistent with previous observations (e.g. Figs. 3 and 4).
= 100 species with Uniform(50000,350000)-distributed genome sizes and Pareto(1,3.5)-distributed abundances (solid lines) vs. their targeted sizes (dashed). Consistent with previous observations for this case, the actual contig sizes obtained are slightly smaller than the targeted length. The median minimax contig sizes are 3.68, 4.85 and 6.16 (in read lengths, which is 92–103% of the target length), and 95% of all experiments yield contigs of length 3.38, 4.43 and 5.63 from all species (85–94% of the target length). The slight undersizing of contigs is consistent with previous observations (e.g. Figs. 3 and 4).

