Table 1. Designs for viral metagenome experiments.
S ( ) ) |

|
Eq. 3 Reads | Eq. 4 Reads | Eq. 5 Reads | (5,50,95)% minimax |
| 100 | 4 | 62402 | 62166 | 67109 | 3.38, 3.68, 3.94 |
| 200 | 4 | 128399 | 135996 | 142673 | 3.32, 3.60, 3.80 |
| 400 | 4 | 263645 | 303155 | 310081 | 3.24, 3.49, 3.72 |
| 100 | 5 | 88636 | 89303 | 96992 | 4.43, 4.85, 5.15 |
| 200 | 5 | 181745 | 196402 | 206985 | 4.28, 4.70, 5.04 |
| 400 | 5 | 371999 | 438059 | 449314 | 4.23, 4.56, 4.78 |
| 100 | 6 | 113767 | 115738 | 126271 | 5.63, 6.16, 6.50 |
| 200 | 6 | 232749 | 255203 | 269879 | 5.39, 5.92, 6.27 |
| 400 | 6 | 475413 | 569204 | 585113 | 5.17, 5.73, 6.04 |
Table 1 provides the numbers of reads of size 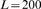 determined to give 95% probability of assembling contigs of at least size
determined to give 95% probability of assembling contigs of at least size  in viral (
in viral ( = 200000,
= 200000,  = Uniform(50000,350000)) metagenomics problems as a function of the number of species
= Uniform(50000,350000)) metagenomics problems as a function of the number of species  or
or  in the pool. Calculations are provided for models using fixed pool and equal genome sizes and abundances (Eq. 3), fixed pool sizes with distributed genome sizes and abundances (Eq. 4) and stochastic pool sizes with distributed genome sizes and abundances (Eq. 5). (5, 50, 95)% minimax contig size quantiles from simulated assemblies of
in the pool. Calculations are provided for models using fixed pool and equal genome sizes and abundances (Eq. 3), fixed pool sizes with distributed genome sizes and abundances (Eq. 4) and stochastic pool sizes with distributed genome sizes and abundances (Eq. 5). (5, 50, 95)% minimax contig size quantiles from simulated assemblies of  species with uniformly distributed genome sizes and Pareto distributed abundances using stochastic pool size/distributed genome size and abundance experimental designs are provided for verification. Larger numbers of reads are required to obtain a given level of performance as pool sizes increase, the required performance level increases, if an assumption of equal genome sizes and abundances is replaced with one of distributed genome sizes/abundances with equivalent mean genome sizes, or if a fixed pool size is replaced with a stochastic pool. Consistent with previous observations, minimax contig size quantiles are slightly (less than one read length) lower than planned.
species with uniformly distributed genome sizes and Pareto distributed abundances using stochastic pool size/distributed genome size and abundance experimental designs are provided for verification. Larger numbers of reads are required to obtain a given level of performance as pool sizes increase, the required performance level increases, if an assumption of equal genome sizes and abundances is replaced with one of distributed genome sizes/abundances with equivalent mean genome sizes, or if a fixed pool size is replaced with a stochastic pool. Consistent with previous observations, minimax contig size quantiles are slightly (less than one read length) lower than planned.
