Table 2. Designs for bacterial metagenome experiments.
S ( ) ) |
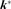
|
Eq. 3 Reads | Eq. 4 Reads | Eq. 5 Reads | (5,50,95)% minimax |
| 100 | 4 | 313476 | 336963 | 365122 | 3.33, 3.49, 3.65 |
| 200 | 4 | 642683 | 766366 | 807394 | 3.27, 3.44, 3.59 |
| 400 | 4 | 1315088 | 1764672 | 1806689 | 3.21, 3.38, 3.51 |
| 100 | 5 | 489834 | 535031 | 584071 | 4.32, 4.51, 4.75 |
| 200 | 5 | 1000506 | 1217621 | 1290028 | 4.17, 4.43, 4.61 |
| 400 | 5 | 2040273 | 2800352 | 2877594 | 4.13, 4.37, 4.59 |
| 100 | 6 | 669257 | 739677 | 811931 | 5.26, 5.63, 5.92 |
| 200 | 6 | 1363646 | 1683594 | 1791344 | 5.20, 5.57, 5.83 |
| 400 | 6 | 2774570 | 3868329 | 3986651 | 5.06, 5.36, 5.59 |
Table 2 provides the numbers of reads of size  determined to give 95% probability of assembling contigs of at least size
determined to give 95% probability of assembling contigs of at least size  in bacterial (
in bacterial ( = 2000000,
= 2000000,  = Uniform(1000000,3000000)) metagenomics problems as a function of the number of species
= Uniform(1000000,3000000)) metagenomics problems as a function of the number of species  or
or  in the pool. Calculations and relationships between both experimental terms and the required number of reads and planned and observed contig sizes are as described in Table 1.
in the pool. Calculations and relationships between both experimental terms and the required number of reads and planned and observed contig sizes are as described in Table 1.
