Table 2. Unjustified annotation percentage of validated problematic domains in protein information resource (PIR) iproclass v3.74 (Global-mode search).
| Domain Name | Type, validated region of model (size) | No. of retrieved sequences | No. of FP hits where 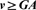 , , 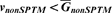
|
No. of annotations without hmmpfam hits (E>10) | Total No. of unjustified hits (%) |
PF00690.18 : Cation_ATPase_N (Cation transporter/ATPase, N-terminus), 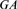 = 18.90, = 18.90, 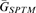 = 9.58, = 9.58, 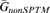 = 18.79, = 18.79,  = −9.47, = −9.47, 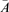 = −76.19 = −76.19 |
TM,66–87 (87), ref.[111] | 3684 | 74 | 3 | 77 (2.1%) |
PF01105.15 : EMP24_GP25L (Endoplasmic reticulum and golgi apparatus trafficking proteins), 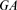 = −16.00, = −16.00, 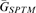 = 13.82, = 13.82, 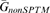 = −20.28, = −20.28,  = −9.54, = −9.54, 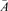 = −208.58 = −208.58 |
TM,141–167 (167), ref. [53] | 1029 | 8 | 33 | 41 (4.0%) |
PF01299.9 : Lamp (Lysosome-associated membrane glycoprotein), 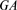 = −87, = −87, 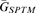 = 18.34, = 18.34, 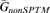 = −95.80, = −95.80,  = −9.54, = −9.54, 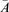 = −614.95 = −614.95 |
TM,304–340 (340), ref. [56] | 164 | 2 | 12 | 14 (8.5%) |
PF01544.10 : CorA (CorA-like Mg2+ transporter protein) 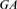 = −61.3, = −61.3, 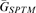 = 28.57, = 28.57, 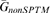 = −80.17, = −80.17,  = −9.70, = −9.70, 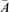 = −503.57 = −503.57 |
TM,341–407 (407), ref. [117] | 2717 | 15 | 71 | 86 (3.2%) |
PF01569.13 : PAP2 (type 2 phosphatidic acid phosphatase) 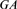 = 8.3, = 8.3,  = 21.70, = 21.70, 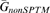 = −3.92, = −3.92,  = −9.47, = −9.47, 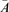 = −120.86 = −120.86 |
TM,102–177 (177), ref. [52] | 5231 | 108 | 19 | 127 (2.4%) |
PF02416.8 : MttA_Hcf106 (sec-independent translocation mechanism protein) 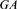 = 7, = 7,  = 17.88, = 17.88, 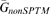 = −1.30, = −1.30,  = −9.58, = −9.58, 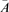 = −102.29 = −102.29 |
TM,1–19 (74), refs. [57], [118] | 2085 | 283 | 0 | 283 (13.6%) |
PF04387.6 : PTPLA (protein tyrosine phosphatase-like protein), 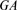 = 25, = 25, 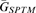 = 13.59, = 13.59, 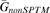 = 20.97, = 20.97,  = −9.56, = −9.56, 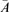 = −291.27 = −291.27 |
TM,89–168 (168), refs. [54], [55] | 277 | 3 | 3 | 6 (2.2%) |
PF04612.4 : Gsp_M (General secretion pathway, M protein) 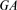 = 25, = 25,  = 24.68, = 24.68, 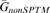 = 10.16, = 10.16,  = −9.85, = −9.85, 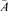 = −247.83 = −247.83 |
TM,1–40 (165), ref. [119] | 401 | 19 | 6 | 25 (6.2%) |
PF07127.3 : GRP (plant glycine rich proteins) 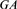 = 17.2, = 17.2, 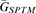 = 14.64, = 14.64,  = 12.16, = 12.16,  = −9.59, = −9.59, 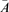 = −173.44 = −173.44 |
SP,1–49 (134), ref. [60] | 207 | 12 | 4 | 16 (7.7%) |
PF08294.3 : TIM21 (Mitochondrial import protein), 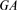 = −20.3, = −20.3,  = 0.19, = 0.19, 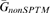 = −10.88, = −10.88,  = −9.61, = −9.61, 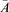 = −309.20 = −309.20 |
TM,1–36 (157), ref. [120] | 118 | 7 | 1 | 8 (6.8%) |
PF08510.4 : PIG-P (phosphatidylinositol N-acetyl-glucosaminyl transferase subunit P), 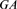 = −11.4, = −11.4, 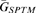 = 40.20, = 40.20, 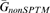 = −42.07, = −42.07,  = −9.53, = −9.53, 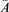 = −233.36 = −233.36 |
TM,1–67 (153), ref. [50] | 143 | 4 | 0 | 4 (2.8%) |
In the first column, we list selected Pfam domains with their accession, identifier, description and their gathering score (as in Pfam release 23) that have TM and/or SP regions included into the model. The region in the domain alignment that includes the validated SP/TM segments (together with interlinking loops as described in Methods) and the corresponding references are provided in the second column. The number of retrieved sequences from iProClass v3.74 with respect to each domain is given in the third column. The number of unjustified hits that returns results (and also satisfied the criteria) and without results are given in the next two columns. The last column gives the total and percentage of the unjustified hits with respect to the number of retrieved sequences. In addition, the log odd scores were re-derived from the match/insert/state transition scores provided by the respective HMM model. The reproduced scores  varied from the original scores at 0.57±0.34.
varied from the original scores at 0.57±0.34. 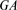 and
and 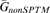 (see equations 19 and 20) denote the domain gathering score threshold and the expected non-SP/TM-specific gathering score threshold respectively.
(see equations 19 and 20) denote the domain gathering score threshold and the expected non-SP/TM-specific gathering score threshold respectively.
Additional material such as hmmpfam outputs and alignments are available at the associated BII WWW site for this work.
