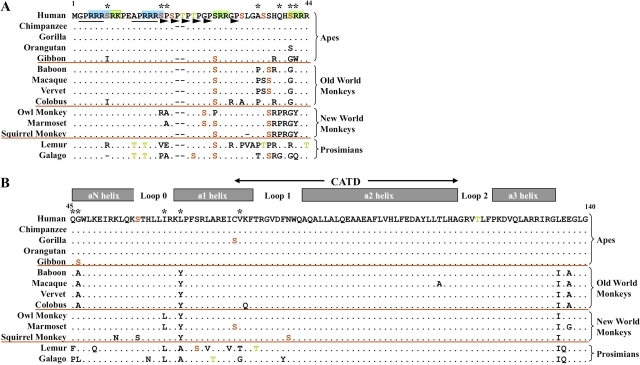
FIG. 1.
Alignment of the deduced CENP-A protein sequences from 14 primates. (A) N-terminal tail region of CENP-A (positions 1–44; residue numbers are based on the human CENP-A sequence). Deviations from the human sequence are indicated by a different amino acid (single-letter abbreviations are depicted); a dot indicates that the amino acid is the same as in the human sequence, and a dash reflects insertion/deletion of an amino acid. Ser, Thr, and Tyr residues predicted to be phosphorylated (table 3 and supplementary table 2, Supplementary Material online) are shown in red, green, and blue, respectively. Deviations from the predicted phosphorylation status of a conserved amino acid are indicated by replacing the black dot by the single-letter amino-acid symbol of the conserved residue in black. Colored letters within the body of the alignment indicate residues predicted to be phosphorylated in that species but not in others. Black asterisks along the top indicate residues under positive selection. The green and blue boxes highlight protein kinase C motifs and cAMP- or cGMP-dependent protein kinase motifs, respectively. SPKK motifs and dipeptide motifs observed in other CENP-A homologs (Malik et al. 2002) are underlined in black or indicated by arrowheads, respectively. Red horizontal lines separate species in each of the four major branches of the primate phylogenetic tree. (B) Histone-fold domain of CENP-A (positions 45–140). Predicted protein structural features (Regnier et al. 2003) are indicated along the top; the CENP-A-targeting domain (CATD; Black et al. 2004) is also indicated. Other features of the alignment are as indicated in A.