Fig. 6.
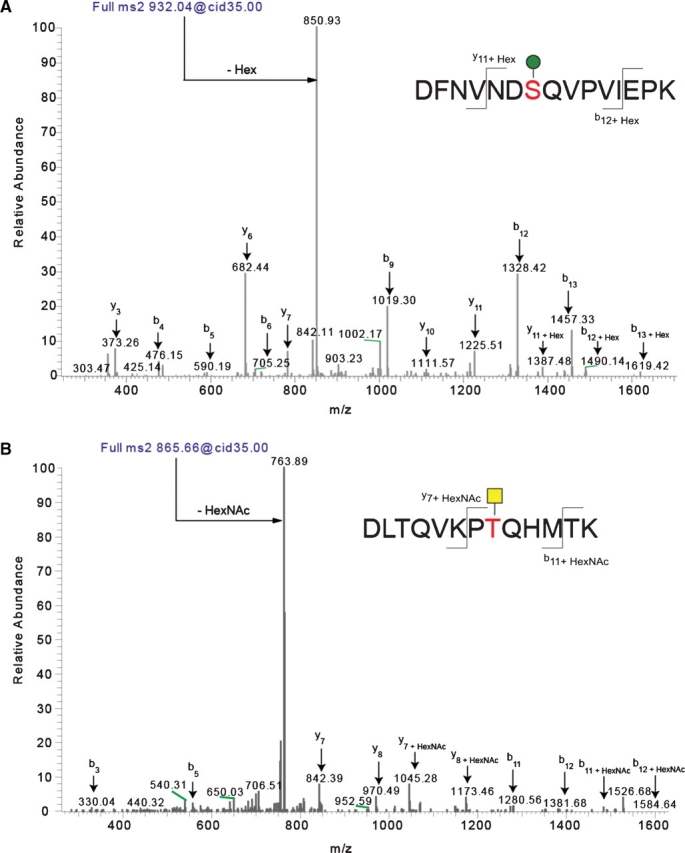
Identification of O-linked sites on ExDG glycopeptides using mass spectrometry. The pseudo-neutral loss method was applied using LC-MS/MS allowing for data-dependent MS3 fragmentation upon witnessing a neutral loss of a glycan mass in the MS2 spectra. Through the application of the pseudo-neutral loss method, it was possible to confirm the assignment of the glycosylated peptide that was identified using TurboSequest. Panel A shows the MS2 of an O-mannose-modified peptide, while panel B shows the MS2 of an O-GalNAc-modified peptide. Note that in both cases the major fragment is the loss of the glycan. Using the MS2 profile (shown) and the MS3 profile of the neutral-loss species (the unmodified peptide, not shown) allowed us to assign the peptide, the glycan, and the site of modification in an automated fashion using TurboSequest followed by manual validation. Sites of modification were manually validated by looking for the addition of Hex and HexNAc to the theoretical m/z of b- and y-ions for the glycopeptide.
