ABSTRACT
Human vaccines, with their exquisite antigenic specificity, have greatly helped to eliminate or dramatically abate the incidence of a number of historical and current plagues, from smallpox to bacterial meningitis. Nonetheless, as new infectious agents emerge and the number of vaccine-preventable diseases increases, the practice and benefits of single-pathogen- or disease-targeted vaccination may be put at risk by constraints of timely production, formulation complexity, and regulatory hurdles. During the last influenza pandemic, extraordinary efforts by vaccine producers and health authorities have had little or no influence on disease prevention or mitigation. Recent research demonstrating the possibility of protecting against all influenza A virus types or even phylogenetically distant pathogens with vaccines based on highly conserved peptide or saccharide sequences is changing our paradigm. “Universal vaccine” strategies could be particularly advantageous to address protection from antibiotic-resistant bacteria and fungi for which no vaccine is currently available.
CURRENT VACCINES: MERITS AND CONSTRAINTS
The current vaccination strategy or “dogma” (1) is that vaccines prepared to fight a given disease are made by one or a few specific antigens of the causative microbial agent or its microbial or viral body with its whole set of antigens. In a few cases, the vaccine is composed of antigenically related strains belonging to the same bacterial species or viral family, such as, for instance, the antituberculous Mycobacterium bovis BCG and smallpox vaccines. When many different types or clades of the same species can cause disease (as is, for instance, the case for pneumonia, bacteremia, and meningitis caused by Streptococcus pneumoniae), the vaccine may be composed of an unusually high number of antigens representative of the most prevalent types or clades (up to 23 polysaccharides in the case of the adult vaccine).
There is nothing more to say about these highly specific vaccines, focused upon a single pathogen or disease, than to acknowledge their extraordinary merits for the preservation of the health of populations. Just to cite a few examples, they have helped to eradicate, eliminate, or control a number of plagues, from smallpox and polio among the viral illnesses to diphtheria and bacterial meningitis among the bacterial illnesses. Together with hygienic water, nothing has probably been more important in the history of infectious diseases and medicine in general than these vaccines, particularly in consideration of their benefits versus their costs. This recognition has led to an ever-increasing appreciation of the medical and social value of vaccination, thus fostering the generation of new vaccines that could substantially broaden the spectrum of vaccine-preventable diseases. Nonetheless, as the number of vaccines increases and old and new diseases join the line for a new vaccine, the practice and benefits of vaccination are being challenged by several factors that, if taken complacently, could severely undermine the confidence in the health benefits of vaccination.
When numerous vaccines are used separately, their acceptability by the population decreases, as they require an increase in the number of visits, administrations, side effects, and costs. Furthermore, when a new vaccine is approved, its insertion into the established vaccination schedule without affecting compliance with vaccination may become problematic. The combination of many different vaccines in a single vial can overcome these issues and has indeed been successfully achieved for some pediatric vaccines. Nonetheless, this procedure raises concerns about the preservation of vaccine quality, long-term stability, and avoidance of negative interactions among the individual antigens. In addition, multidose vaccines usually require blending with a preservative to keep their stability and avoid contamination. The most-used preservative, thimerosal (Merthiolate), continues to be publicly debated for its potential side effects on the nervous system, despite scientific evidence to the contrary. Finally, combination vaccines may not be suitable for poor countries and may not be able to keep up with rapid epidemiological changes. Overall, it is hard to imagine that putting all of the needed vaccines together in a single vial can be a solution for the future.
Particular problems arise when the vaccine target is a microbe or a virus spread in nature as multiple variants (serogroups, serotypes, clades, etc.), all causing the same infections. This is a challenge, for instance, with both the meningococci and still more with pneumococci (2). In the case of Streptococcus pneumoniae, 91 capsular serotype variants are known, and although most of them are infrequent causes of disease in humans, the number of infectious types remains high. The 23-polysaccharide vaccine generates opsonic antibodies directed against serotype-specific capsular polysaccharides and is safe and protective but does not immunize <2-year-old children (an age range that includes a high proportion of the most severe cases of pneumococcal bacteremia and meningitis). In addition, its efficacy in immunocompromised patients is quite inconsistent. Glycoconjugate vaccines have been generated with the capsular polysaccharides of the most widespread and aggressive S. pneumoniae serotypes. They induce T-cell-dependent immunity and opsonizing antibodies directed against type-specific capsular polysaccharides even in very young children (2). These vaccines are also safe and highly protective against invasive S. pneumoniae infections, but serotype shifting may elude the antibody response and displace the vaccine strain. A major event of this kind has been the replacement of vaccine serotypes with nonvaccine serotype 19A, a serotype of S. pneumoniae that is prevalent worldwide, is clinically important, and has the potential for multidrug resistance (2, 3). Thus, we are in a continuous and breathless race, chasing an elusive and dreadful threat with the best of our current glycoconjugate technology to increase the number of vaccine constituents and ensure an efficient formulation. However successful, all of this requires cumbersome and costly reformulation of the vaccine from time to time, with an obvious upper limit.
Fortunately, pneumococcal vaccines do not change every year, as influenza vaccine does! The latter probably constitutes an extreme example of the limitations posed by vaccines with highly focused specificity. Owing to the high variability of its main antigenic constituents inducing neutralizing antibodies (i.e., hemagglutinin [HA] and neuraminidase, glycoproteins of the viral envelope), annual reformulation of a previous vaccine or simply the generation of a totally new one is needed to achieve sufficient protection of the population against the changing threat. Vaccine production, testing for effectiveness, approval by regulatory bodies, distribution, and use require something approaching 1 year, a time frame that can make the vaccine useless. The severe limitations of this situation are most acutely apparent when a new pandemic virus, spreading globally in few weeks, emerges: in the last three influenza pandemics (1957, 1968, and the 2009 ongoing one), when a vaccine could be and was indeed produced, the impact, if any, of vaccination on disease prevention or mitigation has been low (4; http://www.who.int). This is particularly frustrating when, as in the last influenza A (H1N1) 2009 pandemic, the occurrence of a new pandemic had long been anticipated, sustained efforts by public health authorities and international scientific organizations were brought to the highest possible preparedness level, and vaccine manufacturers were forced to make a major, unprecedented technological and resourceful effort to produce a vaccine as rapidly as possible.
INFLUENZA: RUSHING TO UNIVERSALITY
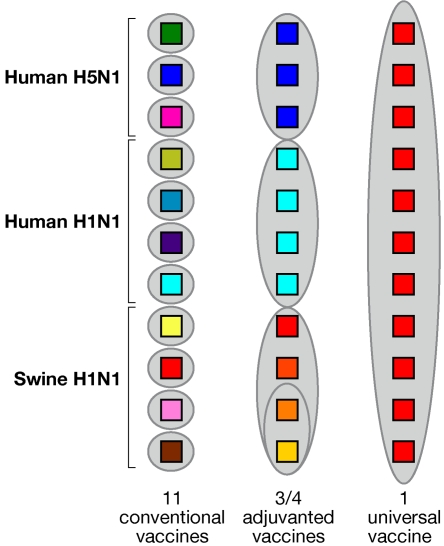
To confront these remarkable problems, various options and technological advances for the more rapid production of vaccines are being pursued, and these will clearly help. Nonetheless, it is unlikely that the challenge will be met exclusively by technology, since this also requires an evolution of our approach to vaccine generation aimed at identifying commonalities in a world of antigenic diversity. With specific reference to influenza, it has long been suspected that a solution could come from the identification of highly conserved sequences in the viral genome and the construction of a vaccine accordingly. In this launching issue of mBio, Steel et al. (5) provide an example of the application of the above-described concepts and technical advances by demonstrating that a vaccine based on a highly conserved sequence of the HA stalk region elicited cross-protective antibodies and was broadly protective in a murine influenza model. This follows a number of older and more recent contributions identifying human antibodies generated by immunization with inactivated seasonal influenza vaccine or just screened from combinatorial antibody libraries, which were cross-reactive with such distant H1 and H5 HAs. These monoclonal antibodies bound to a conserved epitope of the nonglobular portion of the HA molecule close to or containing the fusion peptide (6). Previously, other regions of the influenza virus, pertaining to either the nucleoprotein or the M2 protein of the pericapsidic virus membrane, had been identified; these regions contain highly conserved sequences suitable, in principle, for the generation of cross-protective antibodies and cell-mediated immunity. This may occur especially when the antigenic construct is linked to ligands of Toll-like receptors for efficient stimulation of innate immunity (7–10). In this regard, it is of interest that better coverage of influenza can be obtained by the use of oil-in-water adjuvants, such as MF59 and ASO3, which have turned out to be potent stimulators of innate immunity (11). In a recent work, it has been shown that MF59 may render highly immunodominant also those epitopes which are of low intrinsic dominance when other adjuvants are used (12), thus helping to induce strong specific antibody responses. Figure 1 schematically summarizes the path of influenza vaccine progress. Traditionally, we have had to use a different vaccine for every single virus variant; however, today we can protect against a subgroup of strains by using an adjuvanted vaccine able to cover the diversity of closely related viruses. It is hoped that, in the near future, universal vaccines will be the final solution to pandemic and seasonal influenza.
FIG 1.
Schematic representation of the progress in the development of vaccines against the most recent pandemic and seasonal H1N1 influenza virus strains. The use of adjuvants already allows the coverage of closely related strains with one vaccine. In the future, a universal vaccine may cover all strains.
RESTRICTED UNIVERSALITY: A PRACTICAL SOLUTION FOR VACCINES AGAINST A DEFINED SPECIES OR GROUP OF PATHOGENS
Is influenza the only disease that warrants approaches for universal vaccines? Clearly, it is not, as the call to extend this vaccination practice to all diseases that need it justifies many other instances where approaches to universal vaccines, meaning sustained efforts to identify common antigenic determinants of types or clades and generate vaccines based on these commonalities, are pursued. Nowadays, the application of this “universality” strategy to vaccines for many other diseases has come out of the clouds of pure empiricism and has been made realistic by the enormous progress made in genomic research, particularly by whole-genome sequencing, reverse genetics, and vaccinology and the use of combinatorial antibody libraries and recombinant DNA technology. The ability to sequence the genomes of microorganisms has been a quantum leap in the ability to mine the microbial blueprint and discover conserved antigens that could not be identified by other technologies. Contributory is also the extensive use of more classic biochemical techniques, particularly with polysaccharides and glycoconjugate technologies, with all of this blended with the extraordinary advances in the knowledge of receptors, ligands, and mechanisms of innate and adaptive immunity (6, 11).
The case for a universal pneumococcal vaccine has already been discussed. A promising approach is one based on a combination of a few highly conserved pneumococcal proteins, inclusive of the pneumolysin derivatives and some cell surface proteins. Examples of broad serotype protection with these protein-based vaccines have been obtained in experimental models (3). A meningococcal vaccine immunizing against all serogroups, including serogroup B, against which there is no available vaccine, is also being addressed (13). With the aim to overcome the high diversity of HIV, a universal vaccine based on a chimeric protein encompassing the 14 most conserved HIV regions, inserted into efficient viral vectors, has been designed and tested in preclinical models (14). A universal vaccine strategy to fight the heterogeneity of arenavirus causing severe human infections has recently been proposed (15). In all of the cases mentioned above, we are dealing with somewhat “restricted universality,” since the vaccines are intended to protect against defined, closely related members of a species or family of infectious agents.
UNRESTRICTED UNIVERSALITY: SOMETHING OF A DREAM
Other, less restricted instances of “universality” with much broader applications and consequences also deserve careful consideration in view of the encouraging preliminary results. They suggest that the “universality” strategy could be particularly appealing, and feasible, for the generation of a vaccine against the large group of bacteria and fungi for which there is so far no vaccine at all.
A prominent one is made up of bacteria that mostly cause health care-associated infections. There is no vaccine against bacteria such as Staphylococcus aureus, Escherichia coli, Pseudomonas spp., and Acinetobacter baumannii, which together are responsible for the majority of the infections mentioned above, ranging from septicemia to pneumonia and urinary tract infections, with millions of cases worldwide and elevated mortality despite antibiotic use (16). Adding to the threat, these bacteria often convey a wide array of antibiotic resistance traits and some strains of A. baumannii are actually resistant to almost all of the antibiotics in use. Species variants and types of these bacteria and the virulence factors possessed by each of them are so numerous and diverse that it is quite unlikely to be possible to have specific vaccines. Recently, a glycoconjugate of a synthetic, deacetylated beta-(1-6)-linked, N-acetylated oligoglucosamine polysaccharide (poly-N-acetylglucosamine) which is shared by practically all of the bacteria listed above has been shown to induce antibodies that opsonize and kill both S. aureus and E. coli and protect against infections by these two bacteria in reliable experimental models (17).
Special attention should also be focused on fungal infections, the most widespread of which (for instance, aspergillosis, cryptococcosis, and invasive candidiasis) typically occur in the setting of immunocompromised patients. Moreover, species of the genus Candida, the most frequent agents of fungal infections, are human commensals and species of the genus Cryptococcus can establish a latent host infection. Immunoevasion can occur through antigen target restriction, immunodepression, latency, and commensalism, which are all conditions that raise remarkable obstacles to vaccination against each single agent or species (18). The perspective would clearly change if we could make available a vaccine protective against all of the main fungal agents in patients who share almost overlapping risk factors (e.g., neutropenia) and could therefore be vaccinated before they became debilitated and immunocompromised. Our approach to a universal vaccine against opportunistic fungal pathogens relied on the use of laminarin, a beta-glucan from algae, which was conjugated with a genetically detoxified diphtheria toxin and used to immunize and protect from both Candida and Aspergillus fungi (19). The anti-beta-glucan antibodies generated by the above vaccination and monoclonal antibodies sharing that specificity proved to be protective also against Cryptococcus (20). Importantly, all of these antibodies showed a direct inhibitory activity against the three pathogens in the absence of host cells. Directly inhibitory antibodies are uncommon and may have advantages for use in immunocompromised patients. An approach with an even wider purpose is so-called “idiotypic vaccination” (21). This is based on immunization with an antibody directed against a wide-spectrum yeast killer toxin to raise anti-idiotypic antibodies that mimic the fungicidal activity of the killer toxin itself. Further support for these universal vaccine approaches comes from recent investigations showing that a vaccine composed of heat-killed yeast (Saccharomyces cerevisiae) cells is protective against coccidiomycosis, besides candidiasis (22, 27; Cassone et al., unpublished data). Still more remarkable is the approach taken by some researchers to generate protective immunity against both Candida albicans and Staphylococcus aureus (two top-ranking causes of health care-associated infections) by the use of a vaccine based on the Candida adhesin Als3 (23, 24).
CONCLUSIONS, PERSPECTIVES, AND CRITICAL ISSUES
There is now hope, sustained by knowledge and technology, for the generation of broadly protective “universal” vaccines restricted to species or groups of closely related pathogens or even cross-family or -kingdom vaccines. Overall, it is time to address a new strategy for vaccination based on antigenic commonalities for cross-protective vaccine production. Of particular interest is the fact that some highly conserved universal sequences such as those present in cell surface or cell wall polysaccharides such as beta-glucan are well-known “pathogen-associated molecular patterns” (PAMP) which are sensed by a host’s innate immune system, with a cascade of immunologic events leading to the activation of antimicrobial effectors and antigen-presenting cells which ultimately determine the fate of antigen-specific adaptive immunity. In theory, PAMP-based vaccines could suitably merge the two phases of immune responses for optimal anti-infectious protection in a way expressing the immunizing potency of an adjuvant-antigen mixture in the same molecule (8, 25).
Beta-glucan-based fungal vaccines can generate fungicidal or fungus growth-inhibitory antibodies (18, 19). Theoretically, bactericidal antibodies could be raised by immunization with functionally similar, highly conserved PAMP such as, for instance, peptidoglycan fragments, alone or conjugated to a carrier. These antibodies would act as antibiotics, and Polonelli and collaborators have coined the term “antibiobodies” for them (26). These antibodies could be a breakthrough in the therapy of immunocompromised patients.
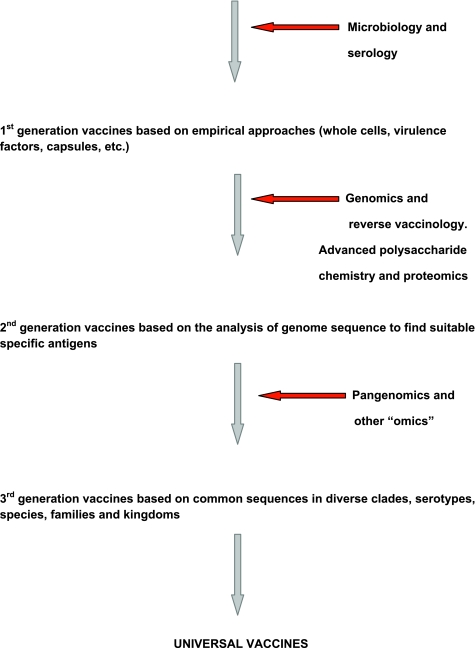
Nonetheless, universal vaccines carry some potential limitations and constraints that must be identified and overcome for their rational exploitation. The first and somewhat obvious one is some defocusing of the immune responses and then a decrease in the capacity to eliminate or keep at bay the etiologic agent. Universal sequences may not be immunodominant, raising the issue of how to potentiate the dominance of antigenic determinants without excessive inflammation. The use of potent viral vectors, presentation as virus-like particles, conjugation with highly immunogenic carriers, and formulation with improved adjuvants such as oil-in-water mixtures or PAMP are some of the tools being exploited. All of the above, in particular the use of PAMP either as an antigen or as a carrier, conveys the possibility of raising autoimmune responses through molecular mimicry or even raising immune responses which dampen the host’s capacity to recognize molecular pattern signatures for a first-line antimicrobial defense. Finally, these broadly specific immune responses might strongly affect the human microbiota, causing excessive elimination of innocent bystanders. Thus, a careful dissection of host beneficial immunity from harmful responses is necessary. Nonetheless, we should not be deterred from in-depth exploration of what is common to a defined type, species, or group of microorganisms, even if they are very distantly related, to move ahead our current strategy of vaccination. Figure 2 schematizes the vaccine history and perspective that are leading those working with vaccines from a merely empirical microbiological stage to a future one which promises to use the best of our “-omics” to generate vaccines using single antigens to protect against many diseases caused by genetically related or even very dissimilar pathogens.
FIG 2.
Schematic view of the history, progress, and perspective of universal vaccines and the accompanying technological tools that make that progress feasible.
ACKNOWLEDGMENTS
We thank Annalisa Pantosti and Antonella Torosantucci (ISS, MIPI Department) for reading the manuscript and helpful discussions.
Footnotes
Citation Cassone, A., and R. Rappuoli. 2010. Universal vaccines: shifting to one for many. mBio 1(1):e00042-10. doi:10.1128/mBio.00042-10.
REFERENCES
- 1. Casadevall A., Pirofski L. A. 2007. Antibody-mediated protection through cross-reactivity introduces a fungal heresy into immunological dogma. Infect. Immun. 75:5074–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan Q. T. 2010. Serious and invasive pediatric pneumococcal disease: epidemiology and vaccine impact in the USA. Expert Rev. Anti Infect. Ther. 8:117–125 [DOI] [PubMed] [Google Scholar]
- 3. Wu K., Zhang X., Shi J., Li N., Li D., Luo M., Cao J., Yin N., Wang H., Xu W., He Y., Yin Y. 2010. Immunization with a combination of three pneumococcal proteins confers additive and broad protection against Streptococcus pneumoniae infections in mice. Infect. Immun. 78:1276–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henderson D. A., Brooke C., Inglesby T. V., Toner E., Nuzzo J. B. 2009. Public health and medical responses to the 1957-58 influenza pandemic. Biosecur. Bioterror. 7:265–273 [DOI] [PubMed] [Google Scholar]
- 5. Steel J., Lowen A. C., Wang T., Yondola M., Gao Q., Haye K., Garcia-Sastre A., Palese P. 2010. An influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1(1):e00018-10 doi:10.1128/mBio.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen G. L., Subbarao K. 2009. Neutralizing antibodies may lead to “universal” vaccine. Nat. Med. 15:1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Du L., Zhou Y., Jiang S. 2010. Research and development of universal influenza vaccines. Microbes Infect. 12:280–286 [DOI] [PubMed] [Google Scholar]
- 8. Roose K., Flers W., Saelens X. 2009. Pandemic preparedness: toward a universal influenza vaccine. Drug News Perspect. 22:80–92 [DOI] [PubMed] [Google Scholar]
- 9. Fiers W., Filette M., El Bakkouri K., Schepens B., Roose K., Schotsaert M., Birkett A., Saelens X. 2009. M2e-based universal influenza A vaccine. Vaccine 27:6280–6283 [DOI] [PubMed] [Google Scholar]
- 10. Huleatt J. W., Nakaar V., Desai P., Huang Y., Hewitt D., Jacobs A., Tang J., McDonald W., Song L., Evans R. K., Umlauf S., Tussey L., Powell T. J. 2008. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 26:201–214 [DOI] [PubMed] [Google Scholar]
- 11. Seib K. L., Dougan G., Rappuoli R. 2010. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet. 5(10):e1000612 doi:1371/journal.pgen.1000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barberà J. P. 2009. MF59-adjuvanted seasonal influenza vaccine. Aging Health 5:475–481 [Google Scholar]
- 13. Holst J. 2007. Strategies for development of universal vaccines against meningococcal serogroup B disease. Hum. Vaccin. 3:290–294 [DOI] [PubMed] [Google Scholar]
- 14. Lètourneau S., Im E. J., Mashishi T., Brereton C., Bridgeman A., Yang H., Dorrell L., Dong T., Korber B., McMichael A. J., Hanke T. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2(10):e984 doi:10.1371/journal.pone.0000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kotturi M. F., Botten J., Sidney J., Bui H. H., Giancola L., Maybeno M., Babin J., Oseroff C., Pasquetto V., Greenbaum J. A., Peters B., Ting J., Do D., Vang L., Alexander J., Grey H., Buchmeierand M. J., Sette A. 2009. A multivalent and cross-protective vaccine strategy against arenaviruses associated with human disease. PLoS Pathog. 5(12):e1000695 doi:10.1371/journal.ppat.1000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson D. J., Kaye K. S. 2009. Controlling antimicrobial resistance in the hospital. Infect. Dis. Clin. North Am. 23:847–864 [DOI] [PubMed] [Google Scholar]
- 17. Gening M. L., Maira-Litràn T., Kropec A., Skurnik D., Grout M., Tsvetkon Y. E., Nifantiev N. E., Pier G. B. 2010. Synthetic β-(1→6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect. Immun. 78:764–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cassone A. 2008. Fungal vaccines: real progress from real challenges. Lancet Infect. Dis. 8:114–124 [DOI] [PubMed] [Google Scholar]
- 19. Torosantucci A., Bromuro C., Chiani P., Bernardis F., Berti F., Galli C., Norelli F., Bellucci C., Polonelli L., Costantino P., Rappuoli R., Cassone A. 2005. A novel glyco-conjugate vaccine against fungal pathogens. J. Exp. Med. 202:597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rachini A., Pietrella D., Lupo P., Torosantucci A., Chiani P., Bromuro C., Proietti C., Bistoni F., Cassone A., Vecchiarelli A. 2007. An anti-β glucan monoclonal antibody inhibits growth and capsule formation of Cryptococcus neoformans in vitro and exerts therapeutic, anti-cryptococcal activity in vivo. Infect. Immun. 75:5085–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Polonelli L., De Bernardis F., Conti S., Boccanera M., Gerloni M., Morace G., Magliani W., Chezzi C., Cassone A. 1994. Idiotypic intravaginal vaccination to protect against candidal vaginitis by secretory yeast, killer toxin-like antiidiotypic antibodies. J. Immunol. 152:3175–3182 [PubMed] [Google Scholar]
- 22. Capilla J., Clemons K. V., Liu M., Levine H. B, Stevens D. A. 2009. Saccharomyces cerevisiae as a vaccine against coccidioidomycosis. Vaccine 27:3662–3668 [DOI] [PubMed] [Google Scholar]
- 23. Lin L., Ibrahim A. S., Xu X., Farber J. M., Avanesian V., Baquir B., Fu Y., French S. W., Edwards J. E., Jr., Spellberg B. 2009. Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog. 5(12):e1000703 doi:10.1371/journal.ppat.1000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baquir B., Lin L., Ibrahim A. S., Fu Y., Avanesian V., Tu A., Edwards J., Jr., Spellberg B. 2010. Immunological reactivity of blood from healthy humans to the rAls3p-N vaccine protein. J. Infect. Dis. 201:473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cassone A., Torosantucci A. 2006. Opportunistic fungi and fungal infections: the challenge of a single, general antifungal vaccine. Expert Rev. Vaccines 5(6):859–867 [DOI] [PubMed] [Google Scholar]
- 26. Polonelli L., Conti S., Gerloni M., Magliani W., Castagnola M., Morace G., Chezzi C. 1991. “Antibiobodies”: antibiotic-like anti-idiotypic antibodies. J. Med. Vet. Mycol. 29:235–242 [DOI] [PubMed] [Google Scholar]
- 27. Stevens D. A. 2010. Abstr. 4th Advances against Aspergillosis Meeting, Rome, Italy, p. 52–53