Fig. 9.
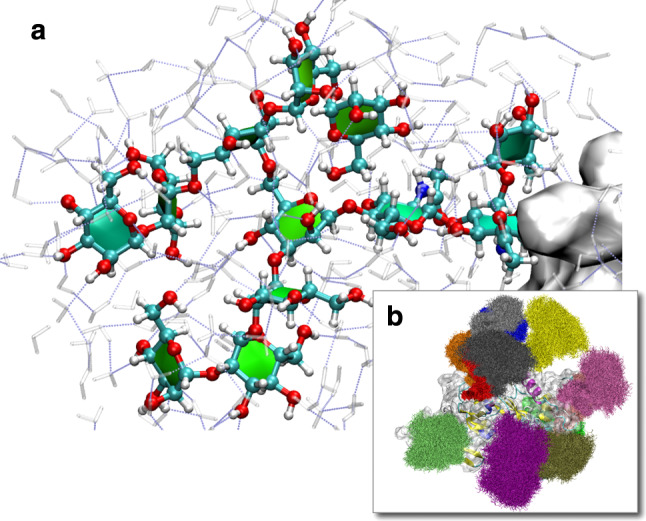
MD simulation of SIV gp120 glycoprotein (M. Frank, unpublished). Complete N-glycans were modeled at 13 glycosylation sites based on the X-ray structure (pdb code 2BF1 [229]). a Solvation shell of a selected N-glycan on the protein surface. b A significant surface area of the protein is shielded by the N-glycans. The molecular system has more than 100,000 atoms (4,832 protein atoms, 3,432 carbohydrate atoms, 30,665 water molecules, 4 chloride ions). Water molecules are not shown for clarity
