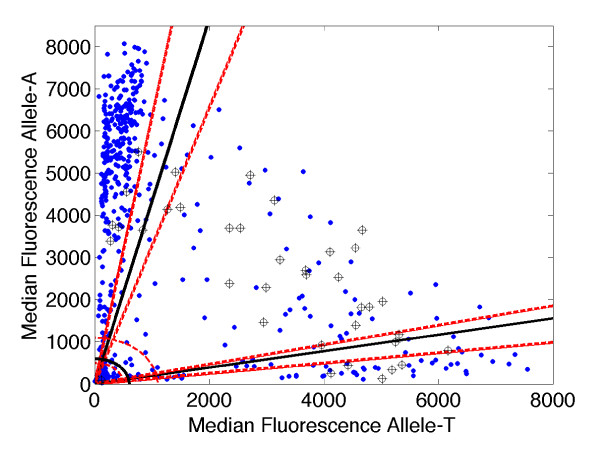
Figure 5.
Control experiment using sample mixtures of known allelic ratios to assess the sensitivity of the histogram segmentation analysis. The LDR-FMA assay was performed for chr1SNP (CombinedSNP.MAL1.1085, PlasmoDB SNP identifier [13]) on 264 samples collected in PNG (blue dots). The polar coordinate histogram method generated diagnostic thresholds (black solid lines) and 95% confidence intervals (red dotted lines). The LDR-FMA assay was performed separately for the same SNP on 34 mixtures of control strains with known genotypes (HB3, allele-T; K1, allele-A) at DNA concentrations ranging from 1:1 to 99:1 (major allele:minor allele, cross-hair symbols) along with a pure sample of each strain and a blank. Samples correctly called as "mixed" had concentration ratios ranging from 1:1 to as high as 15:1. All samples with major:minor allele ratios of 29:1 or higher were misclassified as "single strain infections". Of samples with major:minor allele ratios of 9:1 or smaller, 95% were called correctly (19 of 20); of those with ratios 10:1 or smaller, over 90% were called correctly (22 of 24).