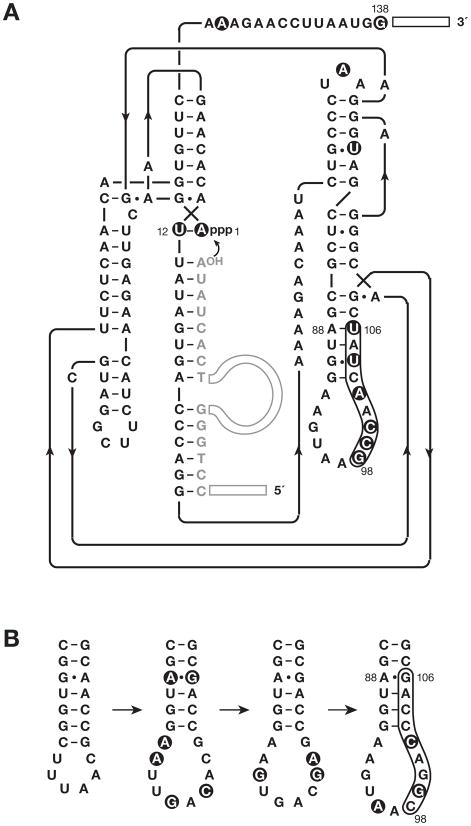
Figure 2.
Sequence and secondary structure of the neomycin-resistant form of the class I ligase. (A) Representative individual from the class of variants that are dependent on neomycin for optimal activity. The secondary structure depiction follows that of the recently reported crystal structure (Shechner et al., 2009). The oligonucleotide substrate is shown in gray. Primer binding sites at the 5′ end of the substrate and 3′ end of the enzyme are depicted as open rectangles. Curved arrow indicates the site of ligation. Filled circles highlight mutations relative to the starting wild-type enzyme. (B) Evolutionary maturation of the P7 stem-loop region, beginning (at left) with the b1-207 form of the enzyme (Ekland et al., 1995), then the evolved variant used to initiate continuous in vitro evolution (Wright and Joyce, 1997), then the E100-3 variant that emerged following 100 successive transfers of continuous evolution (Wright and Joyce, 1997), and finally (at right) the evolved variant that emerged following 500 successive rounds of chip-based continuous evolution (Paegel and Joyce, 2008). The latter variant was used to initiate the present study. Mutations relative to the previous variant in the series are highlighted by filled circles.