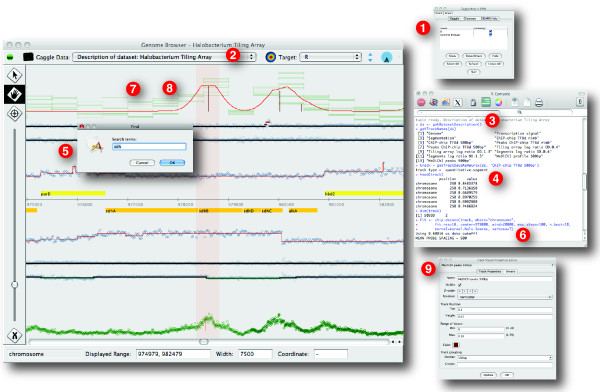
Figure 6.
Processing track data in R with the MeDiChI ChIP-chip deconvolution algorithm. GGB can be used in conjunction with the R environment for statistical computing through the Gaggle framework. After connecting both the genome browser and R to the Gaggle framework (1) we broadcast a description of the dataset using the Gaggle toolbar (2) from the genome browser to R. We can then inspect the dataset in R (3) and load track data into the R environment (4). Locating the region of interest, the sdh operon, using the search feature (5) we then apply MeDiChI's chip.deconv function to the track (6). From R, we broadcast data to the genome browser which then creates new tracks for model fit (7) and peaks (8). We then adjust the visual properties of the new tracks (9) to display predicted transcription factor binding sites.