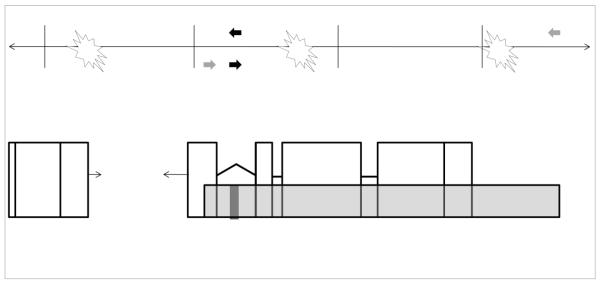
Figure 1. Schematic of the rationale of the QPCR assay for detecting damage in the nuclear genome.
The continuous line ( ↔ ) represents a segment of the nuclear genome and perpendicular hashmarks along the DNA segment mark 10 kb intervals. Open boxes beneath the DNA segment depict genetic coding regions (exons) that are interrupted by interspersed introns (thick black lines). The long amplicon (10-15 kb) is represented as a transparent grey bar that amplifies the majority of a selected gene as well as a portion of the upstream DNA sequence. The bar representing the small amplicon (~200 bases) is shaded black. The filled arrows below and above the DNA segment indicate the position of the corresponding forward and reverse primers for the long (grey) and short (black) amplicon. Lesions are represented as stars that would inhibit or block the progression of the DNA polymerase used in the QPCR assay, thus reducing the amplification of the long product under quantitative conditions. For the smaller amplicon, the relatively close placement of the primers creates a small target size and reduces the probability that such lesions will be detected. Amplification of the short product is not inhibited except by very high levels of damage.