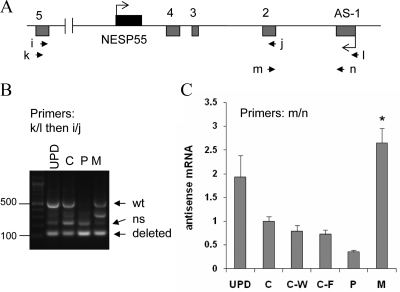
Figure 5.
Amplification of the antisense transcript by RT-PCR using total RNA extracted from LCLs. Panel A, Depiction of GNAS antisense and NESP55 exons. Boxes represent exons; connecting lines, introns; arrowheads, locations of the PCR primers. Panel B, Amplicons generated by nested RT-PCR by using primers k and l followed by primers i and j. Multiple amplicons are due to alternative splicing, as described (6); arrows indicate expected positions of the longest amplicons from the wild-type (wt; 527 bp) and deleted alleles (121 bp). ns, Nonspecific; UPD, patient with patUPD20q; C, healthy normal control; P, unaffected carrier with paternal deletion (individual 3); M, affected individual with maternal deletion (individual 1). Panel C, Real-time RT-PCR results using total RNA from LCLs and primers m and n (mean ± sem of four to six experiments). C-W and C-F are unrelated patients with AD-PHP-Ib, who have STX16 deletions and a loss of exon A/B methylation but show normal differential methylation at the antisense promoter; the methylation status of the antisense promoter was normal in these controls, as reported (28,30,32). Data were normalized to C; *, Value significantly different from C, C-W, and C-F by one-way ANOVA/Tukey’s multiple-comparison test.