Figure 4.
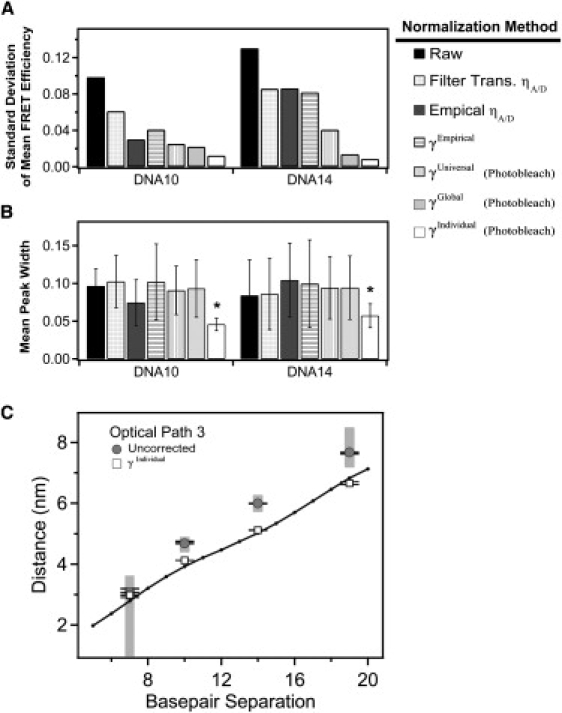
Comparison of methods for normalization of DNA FRET using γPhotobleach. Effectiveness of the normalization methodology was assessed by comparing the mean and width from Gaussian fits to the histogram of a given DNA construct under each of the three different optical paths. (A) The standard deviation in the corrected mean FRET efficiency of each DNA construct measured under the three different optical paths after the indicated method of γ-normalization. Reduced standard deviation indicates convergence of the mean FRET efficiency after γ-normalization is applied. (B) The mean peak width for each DNA duplex under the three optical paths. A reduced mean width indicates narrower peaks after γ-normalization. Error bars indicate the standard deviation between the mean widths measured under different filter sets after the indicated method of γ-normalization. Smaller error bars indicate convergence in the γ-normalized width between the different optical paths. Asterisk denotes statistical significance (p < 0.001), Student's one-tailed t-test. (C) Raw relative proximity ratios (shaded) and γIndividual normalized (open) DNA samples measured under path 3 were converted to distances assuming κ2 = 2/3 and using a Förster's radius of 5.1 nm. The crystal structure of Rhodamine 6G terminally attached to the 5′ end of a DNA double helix (PDB ID: 2V3L) was used as a reference. Using PyMol, distances between the phosphate backbone and the oxygen atom of the central xanthene chromophore were calculated as a function of basepair separation. Error bars indicate the standard deviation for replicate measurements made under the same optical path. The shaded regions correspond to distances calculated from the raw EPR histogram peak widths. In this simple analysis, which does not take into account dye positioning or orientation, γ-normalization results in FRET efficiencies in closer agreement to existing data.
