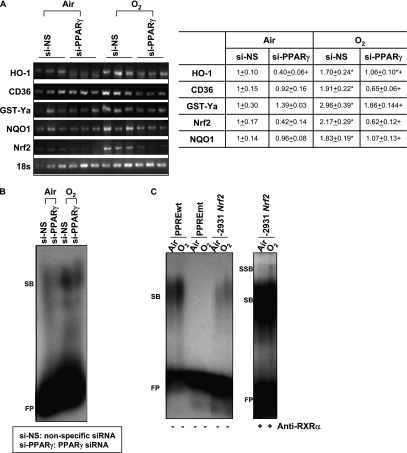
Figure 7.
In vivo peroxisome proliferator activated receptor γ (PPARγ) specific interference RNA (siRNA) treatment suppressed cytoprotective gene expression and antioxidant response elements (ARE) binding activity after hyperoxia. (A) Message levels of cytoprotective genes HO-1, CD36, GST-Ya, and NQO1 and Nrf2 were detected by semiquantitative reverse transcriptase–polymerase chain reaction using total lung RNA isolated from mice treated with nonspecific siRNA (si-NS) or PPARγ-specific siRNA (si-PPARγ) after air and O2 exposure (72 h). cDNA band images for each gene and quantified relative intensities to air-exposed NS-treated mice of digitized cDNA bands normalized to the intensity of each 18s band are shown. Data are presented as group mean ± SEM (n = 3/group). * = significantly higher than treatment-matched air controls (P < 0.05). + = significantly lower than exposure-matched si-NS-treated mice (P < 0.05). (B) Differential nuclear protein-ARE binding activity in the lungs of mice treated with si-NS or si-PPARγ after exposure to air and O2 (72 h). Aliquots of nuclear protein isolated from pooled pieces of left lung tissues (n = 3 mice/group) were incubated with an end-labeled oligonucleotide probe containing ARE consensus sequence. Total ARE binding was determined by gel shift analysis. FP = free ARE probes; SB = shifted bands of total bindings (ARE motif-protein complex). Representative images from multiple analysis (n = 2) are presented. (C) Increased total nuclear protein binding activity (SB) and specific PPARγ-retinoic acid X receptor (RXRa) binding activity (SSB) on an Nrf2 PPAR response element (PPRE)–like sequence in the lungs of mice after exposure to O2 (72 h). Aliquots of nuclear protein isolated from pooled pieces of left lung tissue exposed to either air or O2 (n = 3 mice/group) were incubated with an end-labeled oligonucleotide probe containing -2931 Nrf2 PPRE-like sequence (-2931 Nrf2). PPRE consensus sequence (PPREwt) was used as a positive control for band shift, and mutant PPRE (PPREmt) as a no binding control. FP = free DNA probes; SB = shifted bands of total bindings (PPRE motif-protein complex); SSB = super shifted bands of specific RXRα bindings (PPRE motif-protein–anti-RXRα antibody complex). Representative images from multiple analysis (n = 2) are presented.