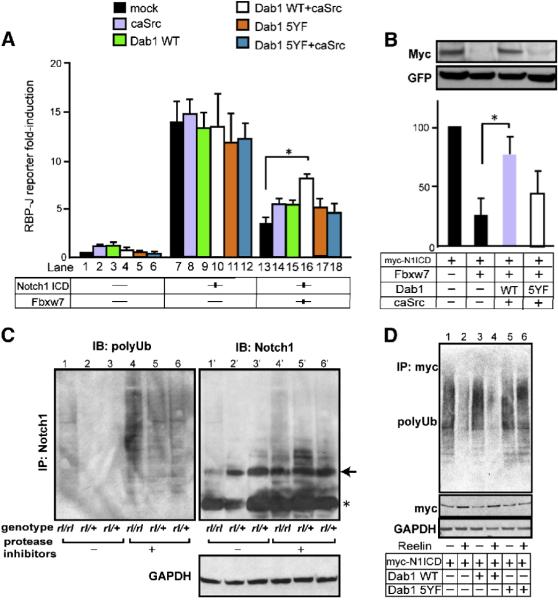
Figure 8. Stimulated Dab1 blocks Notch ICD degradation.
(A) Effects of active Dab1 on Notch ICD activity. Relative Rbpj luciferase activity of each condition is compared to lane 1 set as 1.0. The indicated values are the mean ± SEM of three experiments performed in triplicate. *p<0.05, Student’s t test. (B) Cos-7 cells were transfected with indicated plasmids, and the expression of Notch1 ICD and GFP (as an internal control, reprobed on the same membrane) were analyzed by IB. The graphs show the relative intensity of the Notch1 ICD against the left lane after normalization with GFP as a transfection control. The data represents the mean ± SEM of 3 independent experiments. *p<0.01 paired t-test. Transfection of caSrc without Dab1 or Dab1 without caSrc constructs gave equivalent results to Dab1 5YF with/without caSrc. Transfection of caSrc with/without Dab1 constructs did not alter both endogenous and exogenous Fbxw7 expression levels (data not shown). (C) IB of lysates from rl/rl and rl/+ brain slices against polyubiquitin (FK2) and Notch1 after immunoprecipitation with a Notch1 antibody. IB against GAPDH was obtained from the flow-through of IP. The samples in the left 3 lanes were obtained from freshly prepared cortical slices, and the samples in right 3 lanes were from slices cultured with proteasome inhibitors. Note the decrease of precipitated non-polyubiquitinated Notch1 in Reeler (lanes 1’ and 4’), consistent with the increase of polyubiquitinated Notch1 (lanes 1 and 4). Arrow indicates p100-120 Notch1 ICD bands. Asterisk shows IgG bands. (D) IB of lysates from cultured cortical neurons transfected with the indicated plasmids against polyubiquitin (FK2) and myc after IP with a myc antibody. The transfected neurons were exposed to Reelin containing- or mock medium for 6 hours with protease inhibitors before collection. IB against GAPDH was obtained from the IP flow-through.