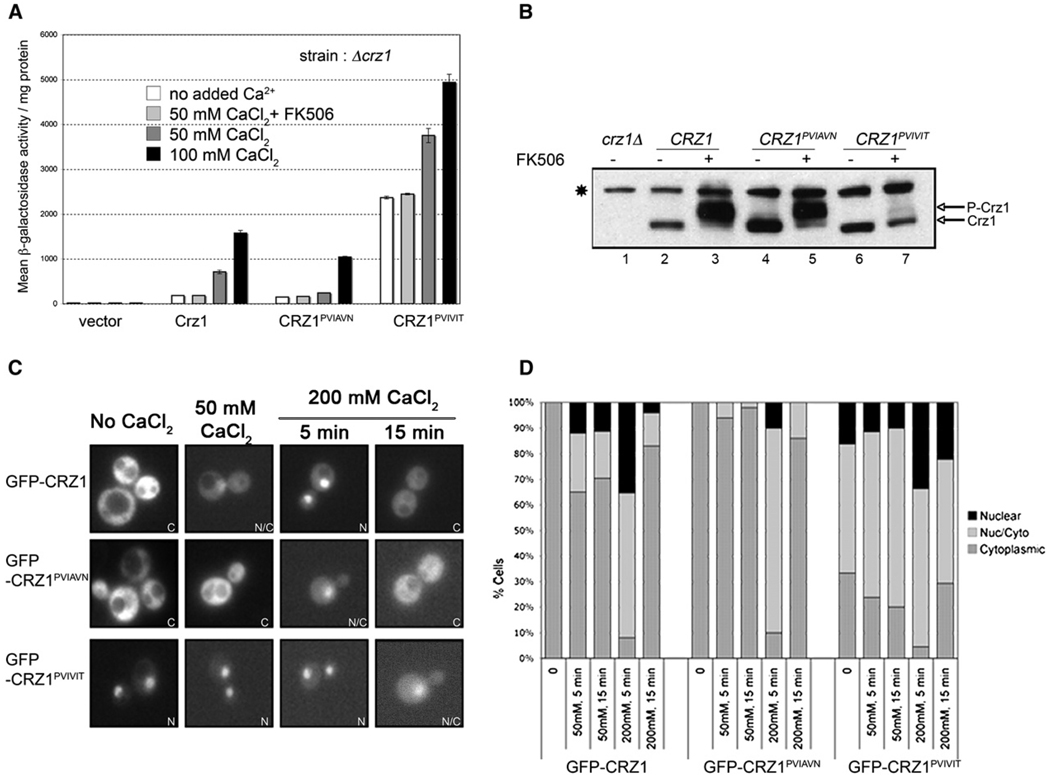
Figure 2. PxIxIT Motif Mutations in Crz1 Alter Transcriptional Activity, In Vivo Phosphorylation States, and Nuclear Localization.
(A) β-galactosidase activity of crzlΔ 4×-CDRE-lacZ strains expressing different Crz1 variants, as noted. All plasmids were constructed in the vector pRS315. CaCl2 was added to the cell culture at the concentrations indicated, 1 hr before harvesting. FK506 was added 1 hr prior to CaCl2, where indicated. “No added Ca2+” refers to cells grown in standard media, which contains 700 µM CaCl2. Error bars indicate standard deviation (see Experimental Procedures for details).
(B) Immunoblot analysis of cell extracts from crz1Δ cells expressing vector or the indicated HA-tagged CRZ1 constructs, using anti-HA monoclonal antibody. Cells were grown either in the presence of FK506 (lanes 3,5, and 7) or solvent (lanes 1,2,4, and 6). CaCl2 (50 mM) was added to the cultures for 1 hr prior to sample preparation. A nonspecific band that is recognized by the anti-HA antibody but is independent of Crz1 is marked with an asterisk (*).
(C) Representative fluorescent images of crz1 Δ strain expressing GFP-Crz1, GFP-Crz1PVIAVN, or GFP-Crz1PVIVIT from pUG36-based plasmids. Cells were visualized within 5–15 min of CaCl2 addition as indicated. “No CaCl2” refers to cells grown in standard media, which contains 700 µM CaCl2. The panels represent examples of the localization category as indicated: N, nuclear; N/C, nuclear/cytoplasmic; and C, cytoplasmic.
(D) Graphical representation of percentage of cells scored as nuclear, nuclear/cytoplasmic, and cytoplasmic from fluorescent images. The data represent the same conditions shown in (C), i.e., no added CaCl2, and 5 or 15 min after addition of 50 mM or 200 mM CaCl2. At least 200 total cells were scored for each condition.