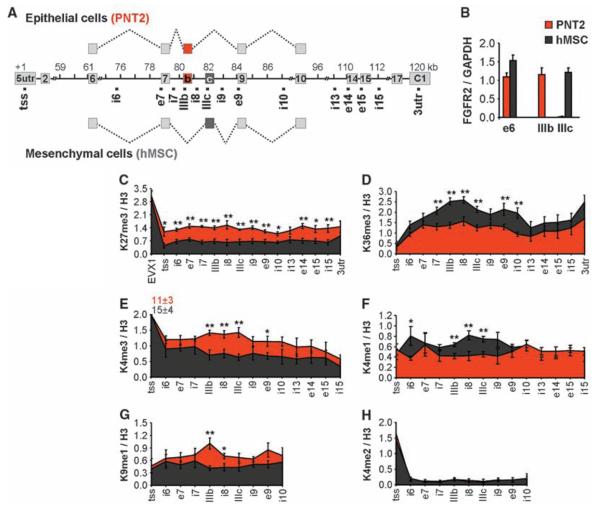
Fig. 1.
Splicing-specific histone modifications. (A) Schematic representation of the human FGFR2 gene. Exon IIIb (red) is included in PNT2 epithelial cells, exon IIIc (black) is included in hMSCs. Square dots indicate oligonucleotide pairs used in analysis. (B) Levels of FGFR2 exon inclusion relative to GAPDH in PNT2 (red) or hMSCs (black) determined by quantitative polymerase chain reaction (PCR). (C to H) Mapping of H3-K27me3 (C), H3-K36me3 (D), H3-K4me3 (E), H3-K4me1 (F), H3-K9me1 (G), and H3-K4me2 (H) in FGFR2 in PNT2 (red) and hMSC (black) cells by quantitative chromatin immunoprecipitation. The percentage of input was normalized to unmodified H3. Values represent means ± SEM from four to six independent experiments. *P < 0.05, **P <0.01, Student's t test.