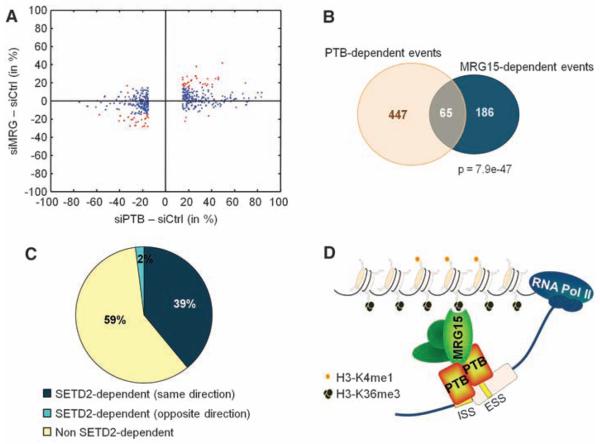
Fig. 5.
Genome-wide identification of SETD2, MRG15, and PTB alternative splicing events in hMSC cells. (A) Scatterplot of AS events in MRG15- (siMRG) or PTB-depleted cells (siPTB) as compared to small interfering RNA control (siCtrl) transfected cells. The cutoff is ≥15% change in exon usage. Red indicates codependent splicing events. (B) Venn diagram of PTB- and MRG15-dependent events with ≥15% change in exon usage. (C) Percentage of MRG15/PTB codependent events that are also sensitive to SETD2 down-regulation. (D) An adaptor system for reading histone marks by the splicing machinery, consisting of a histone mark signature, a chromatin-binding protein (MRG15), and a splicing regulator (PTB).