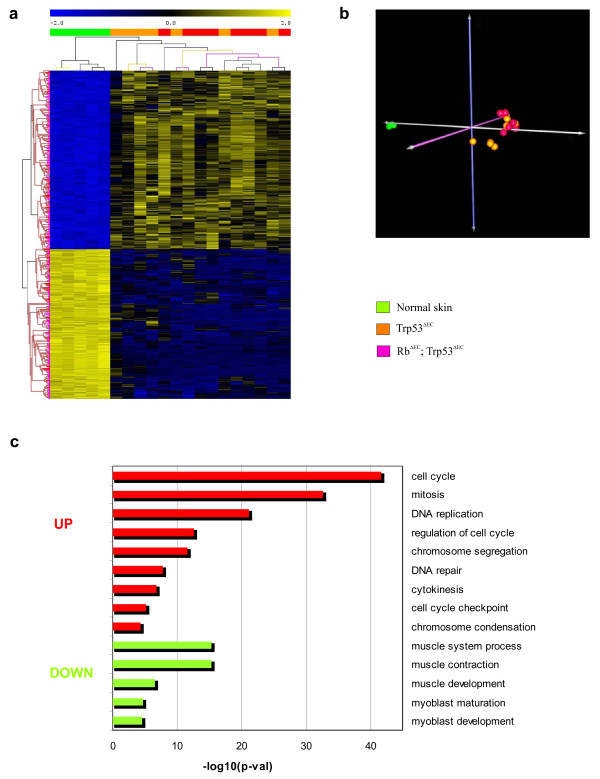
Figure 3.
Tumor signature of p53-deficient mouse. (a) Unsupervised hierarchical clustering of skin tumors using the tumor signature of p53-deficient mouse was done with Pearson distance metrics (average linkage method), and the clustering was iterated 100 times using bootstrap resampling. Columns represent samples, and rows are genes. Green samples are normal control skin from adult mice. Orange samples are skin tumors from Trp53ΔEC genotype, and red samples from RbΔEC; Trp53ΔEC. (b) Principal Component Analysis of animal samples using the tumor signature of p53-deficient mouse. Normal skin and mouse tumors are clearly separated along the principal component 1 (PC1), demonstrating that the signature clearly distinguishes between normal and cancer tissues. Moreover, tumors arising from either genotype (Trp53ΔEC or RbΔEC; Trp53ΔEC) cluster in the same area, showing that the signature is common for them. Green samples are normal skin from adult mice. Orange samples are skin tumors from Trp53ΔEC genotype, and red samples from RbΔEC; Trp53ΔEC. (c) Enrichment analysis in GO biological processes from the gene signature of p53-deficient mouse. Overexpressed genes are related with cell cycle, mitosis, or DNA repair (red bars). Downregulated genes are involved in muscle processes, probably due to loss of muscle tissue in aggressive tumors (green bars). p-val: significance of enrichment.