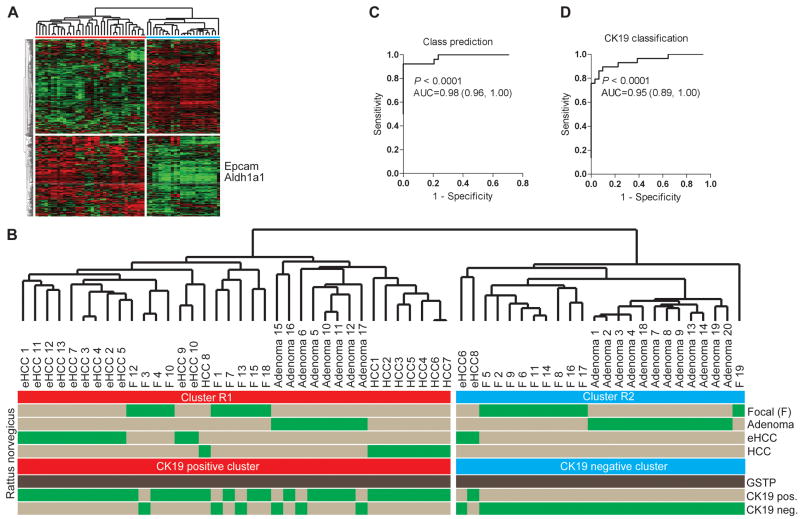
Figure 4.
Development of the CK19-associated gene signature. (A) Unsupervised hierarchical cluster analysis of 60 micro-dissected hepatic lesions including 19 focal lesions at 10 weeks, 20 adenomas at 9 months, 13 early HCCs at 9 months and 8 advanced HCCs at 14 months following DENA administration. A list of 469 differentially expressed genes was computed at P≤0.001 using normal rat liver as a reference. The position of two well characterized progenitor/cancer stem cell markers, Epcam and Aldh1a1, are shown. (B) Unsupervised clustering of the rat lesions. The dendrogram demonstrates a significant separation of the rat lesions according to the CK19 expression profile. The CK19+ lesions belong to cluster R1 (red). (C, D) Class prediction. The probability of correct classification was estimated using a Bayesian compound covariate prediction model for (C) the gene signature (R1 and R2 classification), and (D) the CK19 expression profile during leave-one-out cross-validation (LOOCV). To ensure the accuracy in the prediction method, random permutations were repeated 1,000 times. The degree of the correct classification is represented by the area under the ROC curve (AUC).