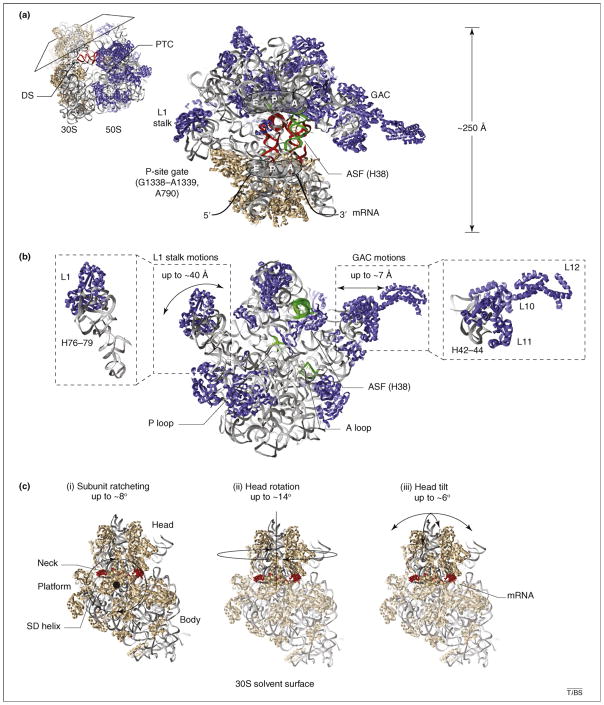
Figure 1.
Structural snapshots of functional ribosome complexes reveal distinct conformational degrees of freedom implicated in the translation mechanism. Distinct views of three-dimensional, atomic models of the bacterial ribosome (Protein Data Bank accession codes 2QAL, 2QAM, 1VS9 and 1Q86) are shown to schematically illustrate (with double-headed arrows) mobile structural elements within (a) the 70S particle, (b) the large (50S) ribosomal subunit and (c) the small (30S) ribosomal subunit. Ribosomal proteins are shown in blue and tan in the large and small subunit, respectively. In both subunits rRNA is shown in gray ribbons. Conserved rRNA elements directly contacting tRNA substrates are shown in green. (a) A cross-sectional view of the 70S pre-translocation ribosome reveals the substrate-binding channel formed at the subunit interface. The three tRNA binding sites (E, P and A) are illustrated, and classically configured A- and P-site tRNAs are shown in red. Landmark structural features, including the decoding site (DS), the peptidyl transferase center (PTC), the L1 stalk, the GTPase activating center (GAC), the A-site finger (ASF; H38 of 23S rRNA), the P-site gate (G1338–A1339, A790 of 16S rRNA) and the mRNA track are indicated for structural reference. (b) The isolated 50S subunit interface is shown to schematically illustrate the protein and rRNA components of the L1 stalk and the GAC and their estimated range of motions [26,55,57]. The ASF bridging the large and small subunit head domains, as well as the A- and P-loop structural elements within the PTC that base pair with the 3′-CCA-ends of tRNA, are highlighted in green. (c) The solvent surface of the 30S subunit is shown to illustrate the single-stranded mRNA (red) track, which wraps around the neck domain and contacts the Shine–Dalgarno (SD) sequence at the convergence of the three principal structural domains (head, platform and body). Panels (i–iii) show the identified conformational degrees of freedom in the ribosome that are implicated in the translocation mechanism and their approximate magnitudes. (i) Subunit ratcheting rotates the three 30S domains counterclockwise in a collective fashion; the reverse motion is termed unratcheting. (ii) Head rotation represents a swiveling motion of the 30S head domain around the neck-like feature connecting the principal domains [3]. (iii) Head tilt represents flexion of the neck perpendicular and parallel to the subunit interface [35]. (ii–iii) The head domain is highlighted to indicate that these motions can largely occur in the absence of subunit ratcheting.