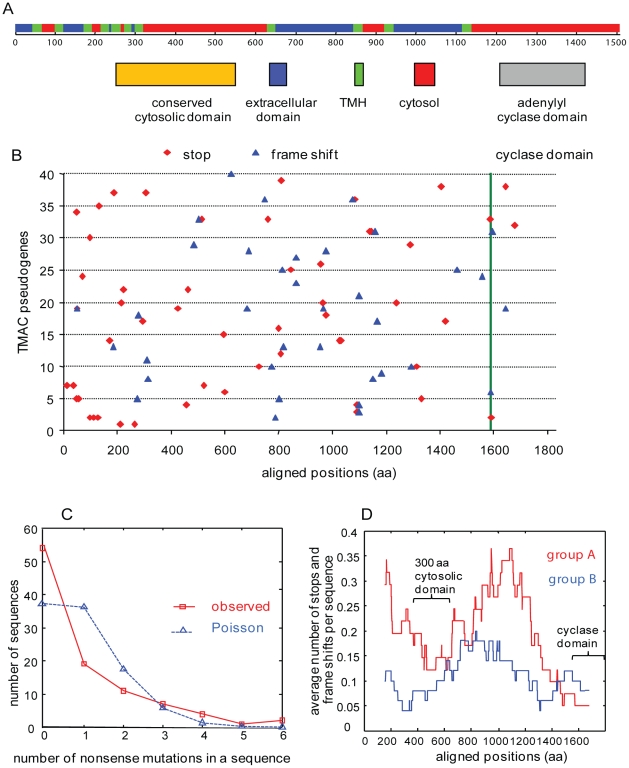
Figure 2. The distribution of stop codons (nonsense mutations) in TMAC genes differs from chance in multiple ways.
A. Each transmembrane adenylyl cyclase (TMAC) has a similar topology predicted by Phobius. There is a series of N-terminal TMHs (green), followed by a conserved cytosolic domain (tan) and a C-terminal cyclase domain (grey) [23]. B. With one exception that may reflect an assembly error, none of the stops and frame shifts is in the same place in 40 aligned TMAC pseudogenes. This result suggests that the stop codons and frame shifts occurred after gene duplication. Note that the alignment technique, which introduces gaps, makes the TMACs appear longer than 1600 amino acids. Green line marks the beginning of the conserved cyclase domain. C. The actual distribution of stop codons among TMAC pseudogenes differs from a Poisson distribution. This result suggests there is selection against the first stop codon (disabling the protein coding capacity of the gene) but not against subsequent stop codons. D. Average number of stops and frame shifts are calculated using a window size of 300 aa in the aligned sequences of group A and B. Stop codons are more abundant in TMAC from the more recently duplicated group A than from group B. Stop codons are also more frequent in less conserved parts of the TMAC genes.