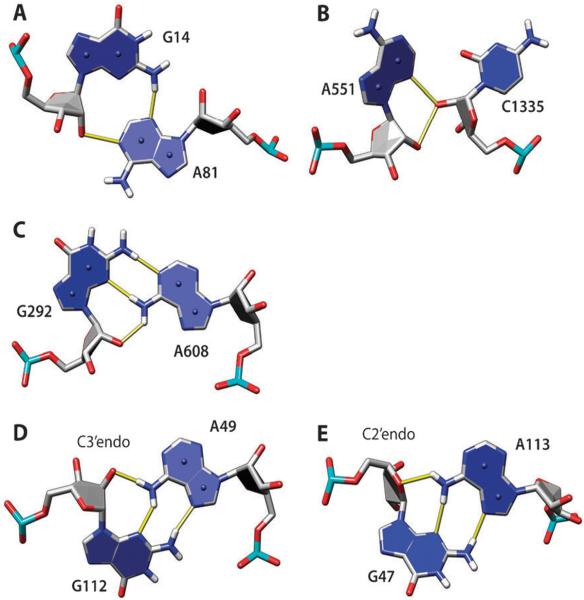
Fig. 2.
Examples of adenine H-bonds with ribose atoms: N1–O2′ (A), N3–O2′ (B), N6–O2′ (C and D), and N6–O4′ (E). The hydroxyl oxygen O2′ serves as an H-bond donor (A, B) or acceptor (C, D). In (A), adenine and guanine make a group 12 AG pair (trans-sugar edge/sugar edge), which is part of the A-minor I motif with G14 Watson–Crick-base paired to C78 and hydroxyl group of C78 making additional H-bonds with the A81 ribose (not shown); G14–C78 is part of a GC-rich helix in the lysine riboswitch (PDB 3DIL). In (B), the unclassified AC pair is part of the A-minor II motif, with A551 interacting with the minor groove of 8-base pair helix (not shown) in 23S rRNA (PDB 1VQ8). In (C), A and G form a group 6 AG pair (trans-Watson–Crick/sugar edge) in 16S rRNA (PDB 2VQE). In (D and E), A and G form group 10 AG pair (trans-Hoogsteen/sugar edge); both are from 23S rRNA (PDB 1VQ8). In (D), the AN1–O2′ H-bond is formed, and in (E), the AN1–O4′ H-bond is formed instead, which is correlated with C2′–endo sugar pucker of G47. Here, and in other molecular graphics, the numbering scheme from the PDB files is used.