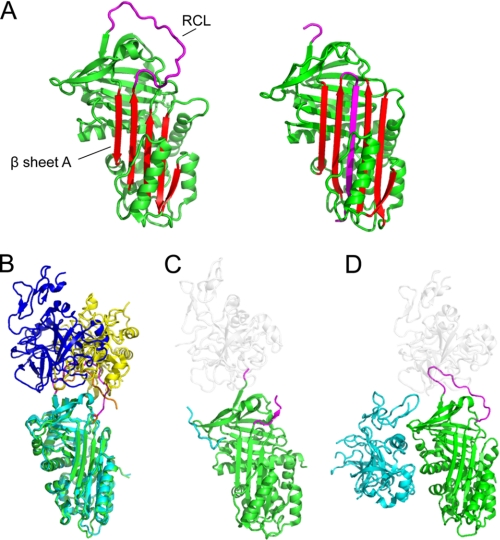
FIGURE 1.
Native serpin structures form docking (Michaelis) complexes with peptidases using exosites and other factors. A, structure of native/stressed (left) and cleaved/relaxed (right) α1AT. β-Sheet A is in red, and the RCL is in magenta. In the cleaved or covalently complexed form of the serpin (see Fig. 2A), the RCL forms an extra strand in sheet A. Native, but not cleaved, serpins are available to form docking complexes. B, superposition of the structure of the antithrombin-thrombin-heparin ternary complex (with serpin in cyan, the RCL in orange, and thrombin in yellow) with antithrombin-fXa-pentasaccharide (with serpin in green, the RCL in magenta, and fXa in blue). The structures are superposed on the serpin. C, structure of α2-antiplasmin (green with magenta RCL). The C-terminal region functions as an exosite for plasmin; the portion visible in electron density is in cyan, and the approximate position of a docking peptidase is shown in ghost white. D, structure of protein Z-dependent inhibitor (green with magenta RCL) in complex with protein Z (cyan). The approximate position of a docking peptidase is in ghost white.