Abstract
Inactivating mutations in hemojuvelin/repulsive guidance molecule c (HJV/RGMc) cause juvenile hemochromatosis (JH), a rapidly progressive iron overload disorder in which expression of hepcidin, a key liver-derived iron-regulatory hormone, is severely diminished. Several growth factors in the bone morphogenetic protein (BMP) family, including BMP2 and BMP6, can stimulate production of hepcidin, a biological effect that may be modified by RGMc. Here we demonstrate that soluble RGMc proteins are potent BMP inhibitors. We find that 50- and 40-kDa RGMc isoforms, when added to cells as highly purified IgG Fc fusion proteins, are able to block the acute effects of both BMP2 and BMP6 at the levels of Smad induction and gene activation, and thus represent a potentially unique class of broad-spectrum BMP antagonists. Whole transcript microarray analysis revealed that BMP2 and BMP6 each stimulated expression of a nearly identical cohort of ∼40 mRNAs in Hep3B cells and demonstrated that 40-kDa RGMc was an effective inhibitor of both growth factors, although its potency was less than that of the known BMP2-selective antagonist, Noggin. We additionally show that JH-linked RGMc mutant proteins that retain the ability to bind BMPs are also able to function as BMP inhibitors, and like the wild type soluble RGMc species, can block BMP-activated hepcidin gene expression. The latter results raise the question of whether disease severity in JH will vary depending on the ability of a given mutant RGMc protein to interact with BMPs.
Keywords: Bone Morphogenetic Protein (BMP), Gene Expression, Genetic Diseases, Iron Metabolism, Protein Chimeras, Protein Purification, Signal Transduction
Introduction
Iron is an essential co-factor for many cellular processes, and plays a vital role in regulating respiration, energy metabolism, and oxygen transport (1, 2). Iron homeostasis is tightly controlled, and detrimental consequences arise from both its deficiency and excess (1, 2). In hemochromatosis, chronic accumulation of excess iron in multiple tissues leads to organ damage and dysfunction (1–3). Juvenile hemochromatosis (JH)2 is a rare and rapidly progressive form of this disorder, and has been linked to alterations in the HJV/HFE2 gene, which encodes for hemojuvelin (HJV) (3, 4). Over two dozen HJV gene mutations have been identified in patients with JH, and include single nucleotide changes that lead to amino acid substitutions and DNA frameshifts that could give rise to truncated proteins (3, 4). In all patients with JH, levels of the key liver-derived iron-regulatory hormone, hepcidin, are reduced, leading to inappropriately elevated iron absorption from the duodenum (3). A similar pattern of excessive iron uptake has been seen in mice engineered to lack HJV (5, 6).
HJV is identical to RGMc, which with RGMa and RGMb comprise the repulsive guidance molecule (RGM) family (4). RGMc is produced in the liver and striated muscle (4, 7, 8), whereas RGMa and RGMb are primarily synthesized in the central nervous system (8), where they are involved in regulating neuronal survival and axonal patterning during development (9, 10). RGM family members are glycoproteins that share several structural motifs (4, 8), and all three can undergo a series of similar biosynthetic and processing steps leading to cell-associated glycosylphosphatidylinositol-linked and soluble protein species (11–13). RGMc is composed of two cell membrane-associated forms, a ∼50-kDa single-chain species and a two-chain isoform with ∼30 and ∼20 kDa disulfide-bonded subunits, and two soluble single-chain members of ∼50 and ∼40 kDa (12, 14). To date, the biochemical mechanisms responsible for production of multiple RGMc protein species have not been elucidated fully.
The precise biological roles for each RGMc species in systemic iron balance also have not been defined yet. Recent studies have hypothesized that cell-associated RGMc functions as a co-receptor for selected bone morphogenetic proteins, and can facilitate the ability of BMPs to stimulate hepcidin gene expression in the liver (15), although evidence supporting this co-receptor concept is incomplete. BMPs are members of the transforming growth factor-β superfamily, and play crucial roles in a variety of developmental and cell fate decisions (16, 17). BMPs bind as dimers to specific type I and type II serine/threonine kinase receptors, and initiate a protein kinase cascade that culminates in the activation by serine phosphorylation of Smads 1, 5, and 8, signal transducers that regulate the expression of many BMP-dependent target genes (16, 17). Several subclasses of BMPs exist that bind to distinct receptors (18, 19). For example, BMP2 and BMP6 are 61% identical in amino acid sequence and belong to the BMP2/4 and BMP5/6/7/8 subclasses, respectively (20). BMP2 preferentially binds to the type II receptor BMPRIIA, and the type I receptor ALK3, whereas BMP6 interacts with ACVRIIa and ALK2 (21). BMP2 also binds with high affinity to the soluble inhibitor, Noggin (22, 23), whereas BMP6 binds poorly (24). Despite these differences, all BMPs promote receptor-mediated activation of Smads 1, 5, and 8, and thus may control the expression of similar groups of genes.
Here we demonstrate that soluble RGMc proteins are potent BMP inhibitors. We find that 50- and 40-kDa RGMc isoforms are able to block the acute effects of both BMP2 and BMP6 at the level of Smad activation, and thus represent a potentially unique class of broad-spectrum BMP modifiers. We additionally show that JH-linked RGMc mutant proteins that retain the ability to bind BMPs also are able to function as BMP inhibitors, and like wild type soluble RGMc species, can block BMP-activated hepcidin gene expression.
EXPERIMENTAL PROCEDURES
Materials
Fetal bovine serum, Ultralow IgG fetal bovine serum, Dulbecco's modified Eagle's medium, modified Eagle's medium, insulin-transferrin-selenium, phosphate-buffered saline, trypsin/EDTA, AcTEV Protease, Protein A-Sepharose 4B, TRIzol reagent, and Superscript III first-strand cDNA synthesis kit were purchased from Invitrogen. N-Glycosidase F (PNGaseF) was from New England Biolabs (Beverly, MA). Ham's F-12 medium was from ThermoScientific Hyclone (Waltham, MA). Recombinant mouse Noggin-Fc, and recombinant human BMP2 and BMP6 were purchased from R&D Systems (Minneapolis, MN). Ni-Sepharose 6 Fast Flow was from GE Healthcare. The furin convertase inhibitor decanoyl-RVKR-CMK and okadaic acid were purchased from Alexis Biochemicals (San Diego, CA); protease inhibitor tablets were from Roche Applied Sciences. Restriction enzymes, buffers, ligases, and polymerases were purchased from Roche Applied Sciences, Clontech, and Fermentas (Hanover, MD). Sodium orthovanadate and dexamethasone were from Sigma. AquaBlock EIA/WIB solution was from East Coast Biologicals (North Berwick, ME). The BCA protein assay kit and GelCode Blue Stain Reagent were purchased from Pierce Biotechnologies. TransIT-LT1 was from Mirus Bio (Madison, WI). NitroBind nitrocellulose was from GE Water & Process Technologies (Trevose, PA). Polyclonal anti-α-tubulin antibody was purchased from Sigma. Monoclonal anti-Smad5 and anti-phospho-Smad5 antibodies were from Abcam (Cambridge, UK). Secondary antibodies, Alexa Fluor 680-conjugated goat anti-rabbit IgG and Alexa Fluor 680-conjugated goat anti-mouse IgG were purchased from Invitrogen. IR800-conjugated goat anti-mouse IgG and IR800-conjugated goat anti-human IgG were from Rockland Immunochemical (Gilbertsville, PA). Other chemicals and reagents were purchased from commercial suppliers.
Cell Culture and Transient Transfections
The following cell lines were purchased from American Type Culture Collection: HEK293 (CRL-1573), Hep3B (HB-8064), and AML12 (CRL-2254). Cells were grown at 37 °C in humidified air and 5% CO2. HEK293 cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal calf serum and transfected at ∼70% confluent density with TransIT-LT1. AML12 cells were maintained in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium with 10% fetal calf serum, 5 μg/ml of insulin, 5 μg/ml of transferrin, 5 ng/ml of selenium, and 40 ng/ml of dexamethasone. Hep3B cells were maintained in modified Eagle's medium plus 10% fetal calf serum.
Generation and Purification of RGMc-IgG1-Fc Fusion Proteins
For generation of full-length RGMc-Fc fusion proteins, codons 1–391 of mouse RGMc were subcloned via 5′ EcoRI and 3′ NotI sites into pcDNA3 (Invitrogen) in-frame with the Fc region of human IgG1 (from pFuse-hFc1, Invivogen, San Diego, CA). Codons comprising a TEV protease recognition site (glutamate-asparagine-lysine-tyrosine-phenylalanine-glutamine), plus three additional glycine residues, were added to the fusion plasmid between RGMc and IgG1-Fc segments, just 5′ to a NotI restriction site. RGMc codon substitutions G92V and G313V, described previously (25), were subcloned into the full-length RGMc-Fc backbone. RGMc truncation mutants were generated by PCR to replace codons after Gln318 and Cys141 with the TEV recognition site and IgG1-Fc. DNA sequencing was used to confirm all nucleotide changes. For generation of IgG1-Fc, codons 1–120 of neogenin (comprising the signal peptide) were subcloned via 5′ HindIII and 3′ NotI sites into pcDNA3 in-frame with the Fc region of IgG1. All RGMc-Fc fusion proteins and IgG1-Fc were purified from conditioned medium of HEK293 cells that were transfected at ∼70% confluent density with 10 μg of DNA per 100-mm diameter culture dish. At 5 h post-transfection medium was changed to Dulbecco's modified Eagle's medium with 2% ultralow IgG fetal bovine serum, and conditioned medium was collected 48 h later. To minimize proteolytic cleavage of RGMc-Fc fusion proteins, the furin convertase inhibitor decanoyl-RVKR-CMK was added to collection medium at a final concentration of 5 μm (25). Protein purifications were performed by IgG affinity chromatography. Conditioned medium was incubated with protein A-Sepharose 4B for 16 h at 4 °C. Bound proteins were eluted with 100 mm glycine, pH 3.0, followed immediately by neutralization with 1 m Tris-Cl, pH 8.0. Purification was assessed after SDS-PAGE by staining of protein bands with GelCode Blue. Protein concentrations were estimated by comparison with bovine serum albumin standards electrophoresed in adjacent lanes.
Purification of 40-kDa RGMc (RGMc40)
RGMc40-Fc purified from conditioned medium was treated with TEV protease (5 units/10 μg RGMc40-Fc) for 3 h at 20 °C in buffer. Samples were then sequentially incubated with Ni-Sepharose for 2 h at 20 °C and Protein A-Sepharose for 2 h at 20 °C to remove TEV and IgG1-Fc, respectively. Purification was assessed and protein concentrations were estimated as described above.
Analysis of RGMc Proteins
To detect the presence of N-linked sugars, purified RGMc fusion proteins were incubated with PNGaseF (500 units) for 16 h at 37 °C according to the supplier's instructions, and products were resolved after SDS-PAGE by staining of proteins with GelCode Blue, and by immunoblotting as described below. To assess the secondary structure of RGMc proteins, circular dichroism (CD) spectroscopy was performed on an AVIV model 215 CD spectrometer maintained at 4 °C. For each protein 3 to 5 spectra were measured from 190 to 260 nm at 0.5-nm intervals for 3 s at each wavelength, using a protein concentration of 0.2 mg/ml and a path length of 0.1 cm, and results were averaged.
Analysis of BMP-mediated Signaling
Confluent Hep3B or AML12 cells were serum starved for 16 h, followed by addition of BMP2 or BMP6 (0–200 ng/ml) for up to 8 h in the absence or presence of various concentrations of purified IgG1-Fc, Noggin-Fc, RGMc40, or RGMc-Fc fusion proteins. Whole cell protein extracts and RNA were isolated and analyzed as described below.
Protein Extraction and Immunoblotting
Whole cell protein lysates were prepared as described (26), and aliquots were stored at −80 °C until use. Protein samples (20 μg/lane) were resolved by SDS-PAGE and transferred to nitrocellulose membranes. After blocking with 50% AquaBlock solution for 1 h at 20 °C, membranes were incubated sequentially with primary and secondary antibodies. Primary antibodies were used at the following dilutions: anti-phospho-Smad5, 1:1000; anti-Smad5, 1:500; and anti-α-tubulin, 1:30,000. Secondary antibodies were used at 1:10,000. Results were visualized and images captured using the LiCoR Odyssey and version 3.0 analysis software.
RNA Isolation and Analysis
Total cellular RNA was extracted using TRIzol reagent. RNA concentrations were determined spectrophotometrically (absorbance at a ratio of 260 to 280 nm (A260/280) ≥ 1.9), and RNA quality was assessed by agarose gel electrophoresis. RNA (2 μg) was reverse transcribed with the Superscript III first-strand cDNA synthesis kit using oligo(dT) primers in a final volume of 25 μl. PCR were performed with 0.5 μl of cDNA according to a protocol supplied with the Advantage2 GC kit (Clontech), using the primer pairs listed in Table 1. PCR assays were performed within the linear range of cycle numbers for each primer pair (18–35). PCR products were separated on 1.2% agarose gels and images were captured and quantified with a GelDoc imager and Quantity One® software (Bio-Rad).
TABLE 1.
Primers used for RT-PCR
| Gene | Location | DNA sequence (5′ – 3′) | cDNA size |
|---|---|---|---|
| bp | |||
| ATOH8 | Exon 1 | CCTCCTCCGAGATCAAAGC | 219 |
| Exon 2 | CGGCACTGTAGTCAAGGTCA | ||
| DUSPI | Exon 3 | CTGCCTTGATCAACGTCTCA | 160 |
| Exon 4 | ACCCTTCCTCCAGCATTCTT | ||
| GATA2 | Exon 2 | GTCACTGACGGAGAGCATGA | 232 |
| Exon 3 | GCCTTCTGAACAGGAACGAG | ||
| HAMP | Exon 1 | TGGCACTGAGCTCCCAGATC | 209 |
| (human hepcidin) | Exon 3 | CGCAGCAGAAAATGCAGATG | |
| ID1 | Exon 1 | AAACGTGCTGCTCTACGACA | 153 |
| Exon 1 | GATTCCGAGTTCAGCTCCAA | ||
| SLC6A19 | Exon 11 | ACCCTGGCTACGAGGAATTT | 212 |
| Exon 12 | GTACTTCAGGTCCCCGTTCA | ||
| SMAD7 | Exon 1 | TCCTGCTGTGCAAAGTGTTC | 211 |
| Exon 2/3 | TCTGGACAGTCTGCAGTTGG | ||
| S17 | Exon 2 | CATTATCCCCAGCAAAAAGC | 155 |
| Exon 3/4 | AGGCTGAGACCTCAGGAACA | ||
| Hamp1 | Exon 2 | GAGACAGACTACAGAGCTGCAG | 182 |
| (mouse hepcidin 1) | Exon 3 | GTCAGGATGTGGCTCTAGGCTA | |
| S17 | Exon 2 | ATCCCCAGCAAGAAGCTTCGGAACA | 332 |
| Exon 5 | TATGGCATAACAGATTAAACAGCTC |
Microarray Analysis of Gene Expression
Hep3B cells were treated with 100 ng/ml of BMP2 or BMP6 alone or in combination with a 10-fold molar excess of Noggin-Fc or a 20-fold molar excess of RGMc40-Fc for 4 h. Untreated Hep3B cells were used as controls. Total cellular RNA was extracted as described above, followed by an additional sodium acetate-ethanol precipitation. Preparation of cDNAs, labeling, hybridization, quality control, and data acquisition were performed at the OHSU Gene Microarray Shared Resource using the Affymetrix GeneChip® Human Gene 1.0 ST Array. Two biological replicates were obtained for each treatment group. Results were normalized using the robust multichip average algorithm and analyzed with web-based GeneSifter® software. Transcripts from log2-transformed data were filtered according to the following criteria: for pairwise comparisons, t test, p < 0.05; and for multigroup comparison analysis, one-way analysis of variance, p < 0.02. For all comparisons the Benjamini and Hochberg correction was applied. The complete microarray data set (series record GSE20671) may be found at the National Center for Biotechnology Information Gene Expression Omnibus.
RESULTS
BMP2 and BMP6 Rapidly and Potently Stimulate Hepcidin Gene Expression in the Hep3B Liver Cell Line
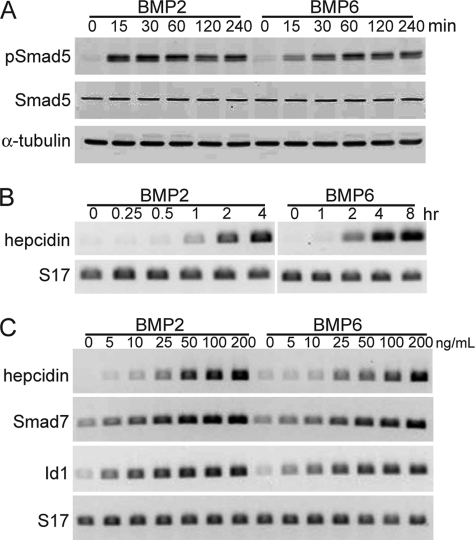
Several BMPs have been linked to regulation of systemic iron metabolism through effects on hepcidin gene expression in the liver (15, 27–29), and it has been proposed that RGMc modifies these actions of BMPs (15, 27, 30, 31), although the biochemical and molecular mechanisms have not been elucidated. Here we have asked if Hep3B cells could provide a good model for studying the interactions of RGMc with different BMPs. Incubation of either BMP2 or BMP6 with confluent Hep3B cells led to rapid stimulation of intracellular signaling via BMP receptors, as seen by inducible serine phosphorylation of Smad5 within 15 min of growth factor addition that was sustained for up to 240 min (Fig. 1A). Both BMPs also rapidly induced hepcidin gene expression, with mRNA being detectable within 1 h after BMP2 treatment, and within 2 h after addition of BMP6 (Fig. 1B). In contrast, only BMP2 could promote hepcidin mRNA accumulation in the mouse AML12 liver cell line (supplemental Fig. S1), making this an unusable model for examining the actions of different BMPs. Dose-response curves showed that BMP2 was slightly more effective than BMP6 in inducing gene expression in Hep3B cells, as indicated by ∼2-fold greater increases in hepcidin, SMAD7, and ID1 mRNAs at any growth factor dose up to 200 ng/ml (Fig. 1C). Taken together, these results show that both BMP2 and BMP6 rapidly activate Smads and stimulate gene expression in Hep3B cells, and indicate that this cell line would be a good model for examining the effects of different RGMc protein species on BMP-mediated signaling and biological effects.
FIGURE 1.
BMP2 and BMP6 rapidly stimulate Smad phosphorylation and gene expression in the Hep3B liver cell line. Human Hep3B cells were incubated in serum-free medium with BMP2 or BMP6 at the indicated doses for up to 8 h, and assayed for activation of Smad5 or induction of gene expression. A, immunoblots of whole cell protein lysates for phosphorylated Smad5 (pSmad5), total Smad5, and α-tubulin after incubation of cells with BMP2 or BMP6 (200 ng/ml) for 0–240 min. B, measurement of hepicidin and S17 mRNAs by RT-PCR after incubation of cells with BMP2 or BMP6 (200 ng/ml) for 0–8 h. C, measurement of hepicidin, SMAD7, ID1, or S17 mRNAs by RT-PCR after incubation of cells with graded doses of BMP2 or BMP6 (0–200 ng/ml) for 4 h. Representative results are shown of 3 independent experiments.
Purification from Mammalian Cells and Biochemical Properties of Recombinant Soluble RGMc Proteins
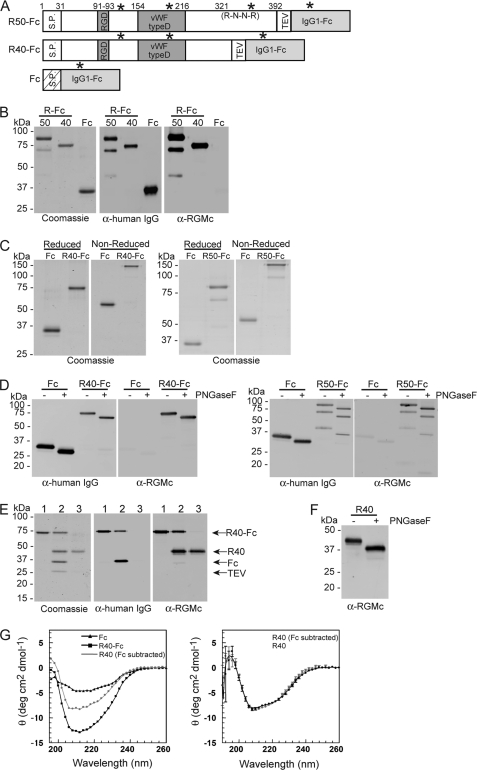
RGMc undergoes a complex series of biochemical and processing steps leading to membrane-bound and extracellular forms of the protein, including ∼50 and ∼40 kDa soluble single-chain species (12, 14, 32, 33) here termed RGMc50 and RGMc40. We expressed both of these isoforms as IgG1-Fc fusion proteins (see Fig. 2A for domain maps) and purified them from HEK293 cell conditioned culture medium by protein A affinity chromatography (Fig. 2B). RGMc40-Fc and IgG1-Fc are predominantly single species of ∼73 and ∼33 kDa, respectively, on reducing SDS-PAGE, as evidenced by staining of the purified proteins with Coomassie Blue, and by detection with antibodies to human IgG and mouse RGMc (Fig. 2B). The expected full-length RGMc50-Fc fusion protein of ∼83 kDa is seen as the major purified band, but two smaller and less abundant immunoreactive species of ∼68 and ∼45 kDa also are detected, and represent cleavage products of the full-length protein.
FIGURE 2.
Purification and characterization of soluble RGMc proteins from mammalian cells. Recombinant mouse RGMc proteins were purified from HEK293 cell culture medium as described under “Experimental Procedures.” A, map of RGMc-IgG1-Fc fusion proteins showing locations of signal peptide (S.P.), RGD sequence, von Willebrand factor type D (vWF type D) domain, pro-protein convertase recognition site (R-N-R-R), TEV protease recognition site, IgG1-Fc domain, and N-linked glycosylation sites (asterisks). R50-Fc and R40-Fc are RGMc50-Fc and RGMc40-Fc, respectively. Also depicted is IgG1-Fc (Fc). B, analysis after reducing SDS-PAGE of R50-Fc, R40-Fc, and Fc by staining with Coomassie Blue (left panel), or after immunoblotting with anti-human IgG antibody (α-human IgG, middle) or anti-RGMc antibody (α-RGMc, right). C, RGMc-Fc fusion proteins are dimers. Coomassie Blue-stained gels showing purified Fc and R40-Fc (left panels) or R50-Fc (right panels) after separation by SDS-PAGE under reducing (left panel) or non-reducing (right panels) conditions. D, R40-Fc, R50-Fc, and Fc are glycosylated. Immunoblots of PNGaseF-treated purified Fc and R40-Fc (left panels) or R50-Fc (right panels) with antibodies to human IgG (left panel) or RGMc (right panel). E, purification of RGMc40 from the Fc fusion protein. Analysis after reducing SDS-PAGE by Coomassie Blue staining (left panel), and immunoblotting with α-human IgG (middle panel) or α-RGMc (right panel). Lane numbers represent: 1, purified R40-Fc; 2, R40-Fc after incubation with TEV for 3 h at 20 °C; 3, purified R40 after sequential Ni-Sepharose and Protein A-Sepharose affinity chromatography (see “Experimental Procedures” for details). F, R40 is glycosylated. An immunoblot of PNGaseF-treated purified R40 with RGMc antibody is shown. For B-F, molecular weight markers are indicated. G, RGMc40 and RGMc40-Fc are structurally similar. Left panel, results of 3 averaged CD spectra for purified RMGc40-Fc (black squares) and IgG1-Fc (black triangles) measured from 190 to 260 nm at 4 °C (left). The gray circles and gray line depict a spectrum for RGMc40 that was calculated by subtracting data for IgG1-Fc from RGMc40-Fc. Right panel, results of 5 averaged CD spectra for purified RGMc40 measured from 190 to 260 nm at 4 °C (black circles) compared with the calculated spectrum from the left panel for RGMc40 (gray circles; corrected for IgG1-Fc). The standard deviation of the measurements is included at every fourth data point.
We performed a series of analytical experiments to evaluate each purified soluble RGMc species. Because IgG proteins naturally form dimers via disulfide linkages through their Fc region (34) we showed that RGMc50-Fc, RGMc40-Fc, and IgG1-Fc exist as dimers by demonstrating that their relative electrophoretic mobility was diminished on non-reducing compared with reducing SDS-PAGE gels (Fig. 2C). RGMc50, RGMc40, and IgG1-Fc each are predicted to contain N-linked sugars (Fig. 2A), and incubation with PNGaseF resulted in enhanced mobility on reducing SDS-PAGE (Fig. 2D). Moreover, after removal of the IgG1-Fc domain by incubation of RGMc40-Fc with TEV protease (Fig. 2E), the resultant purified recombinant RGMc40 was still a substrate for PNGaseF (Fig. 2F), indicating that both the RGMc and IgG1-Fc moieties are N-linked glycoproteins. We analyzed secondary structural characteristics of RGMc40-Fc and RGMc40 by CD spectroscopy, and found that both proteins were very similar to each other in their α-helical and β-sheet content (Fig. 2G). Overall, the results depicted in Fig. 2 demonstrate that both RGMc50-Fc and RGMc40-Fc maintain the characteristics of native RGMc50 and RGMc40 protein species, and indicate that these recombinant soluble RGMc isoforms will be useful tools to probe the functional consequences of interactions with BMPs.
RGMc50 and RGMc40 Inhibit BMP2- and BMP6-mediated Signaling and Gene Activation
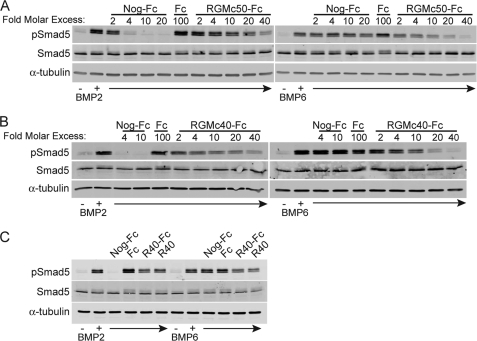
We assessed the effects of RGMc fusion proteins on BMP receptor activation in Hep3B cells by monitoring changes in BMP-stimulated phosphorylation of Smad5. Both RGMc50-Fc and RGMc40-Fc caused a dose-dependent decline in the extent of BMP-mediated Smad5 phosphorylation, with a consistently greater inhibitory effect seen on BMP6 than on BMP2 (Fig. 3, A and B). BMP2 stimulated Smad5 phosphorylation more rapidly than BMP6, and at these earlier time points RGMc40-Fc also effectively inhibited these actions of BMP2 (supplemental Fig. S2). Noggin is a well described inhibitor of BMP2 but is not very effective against BMP6 (22, 23), and Noggin-Fc was more potent than RGMc50-Fc or RGMc40-Fc in blocking the effects of BMP2, as evidenced by a complete inhibition of Smad5 phosphorylation at 4-fold molar excess, but did not reduce the effects of BMP6 at 20-fold molar excess (Fig. 3). In contrast, addition of IgG1-Fc at a dose as high as 100-fold molar excess did not interfere with the actions of either growth factor (Fig. 3). Moreover, as RGMc40-Fc and RGMc40 were equipotent in blocking the effects of BMP2 and BMP6 on Smad5 phosphorylation (Fig. 3C), we conclude that the IgG1-Fc domain does not influence the actions of the RGMc moiety in the recombinant fusion protein.
FIGURE 3.
Dose-dependent inhibition of BMP2- and BMP6-mediated signaling by RGMc50 and RGMc40. BMP2 or BMP6 (100 ng/ml) were preincubated for 3 h at 20 °C with various concentrations of Noggin-Fc (Nog-Fc), IgG1-Fc (Fc), or RGMc proteins, and after addition to cells for 2 h, pSmad5, Smad5, and α-tubulin were detected by immunoblotting. A, Nog-Fc, 2–20-fold molar excess; Fc, 100-fold molar excess; RGMc50-Fc, 2–40-fold molar excess over BMP. B, Nog-Fc, 4–10-fold molar excess; Fc, 100-fold molar excess; RGMc40-Fc, 2–40-fold molar excess over BMP. C, Nog-Fc, 10-fold molar excess; RGMc40-Fc (R40-Fc), 10-fold molar excess; RGMc40 (R40), 10-fold molar excess, or Fc, 100-fold molar excess over BMP.
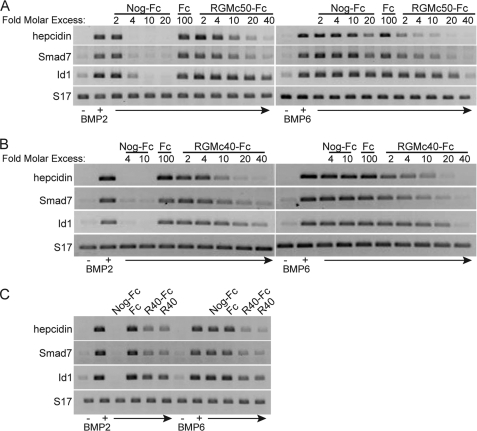
We next examined the impact of RGMc40-Fc and RGMc50-Fc on BMP-activated gene expression. Addition of each fusion protein caused a dose-dependent decrease in the extent of BMP-stimulated accumulation of hepcidin, SMAD7, and ID1 mRNAs, as measured by semi-quantitative RT-PCR, with greater inhibitory effects being observed on BMP6 than on BMP2 (Fig. 4). Of note, ID1 gene expression was reduced less by RGMc-Fc than was hepcidin or SMAD7. As expected, Noggin-Fc was ∼5 times more potent than RGMc-Fc in blocking the actions of BMP2 on gene expression, but had little effect on BMP6, and the IgG1-Fc domain on its own did not interfere with BMP-regulated gene activity at 100-fold molar excess (Fig. 4). In addition, RGMc40 and RGMc40-Fc were equivalently inhibitory (Fig. 4C). Taken together, the results in Figs. 3 and 4 show that RGMc50 and RGMc40 are potentially broad-based inhibitors of BMP-mediated signaling in cultured cells.
FIGURE 4.
Dose-dependent inhibition of BMP2- and BMP6-mediated gene expression by RGMc50 and RGMc40. BMP2 or BMP6 (100 ng/ml) were preincubated for 3 h at 20 °C with various concentrations of Noggin-Fc (Nog-Fc), IgG1-Fc (Fc), or RGMc proteins, and after addition to cells for 4 h, hepcidin, SMAD7, ID1, and S17 mRNA levels were analyzed by RT-PCR. A, Nog-Fc, 2–20-fold molar excess; Fc, 100-fold molar excess; RGMc50-Fc, 2–40-fold molar excess over BMP. B, Nog-Fc, 4–10-fold molar excess; Fc, 100-fold molar excess; RGMc40-Fc, 2–40-fold molar excess over BMP. C, Nog-Fc, 10-fold molar excess; RGMc40-Fc (R40-Fc), 10-fold molar excess; RGMc40 (R40), 10-fold molar excess; Fc, 100-fold molar excess over BMP.
Global Effects of RGMc40 on Gene Expression
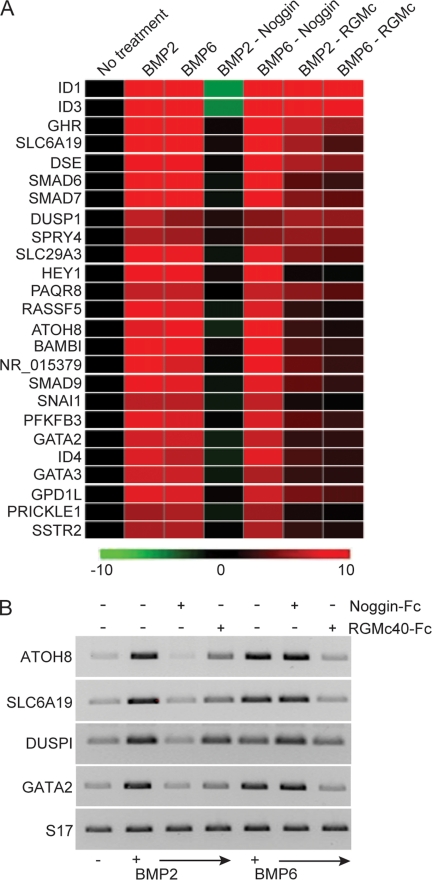
To extend the scope of our experiments on the actions of BMPs and RGMc in Hep3B cells, we examined changes in gene expression using microarray assays. Whole transcript analysis with Human Gene 1.0 ST GeneChip® arrays revealed that BMP2 and BMP6 stimulated the accumulation of a nearly identical cohort of mRNAs, with ∼40 transcripts being induced by at least 2-fold (supplemental Table S1), and 25 being increased by >2.5-fold (Fig. 5A) within 4 h of BMP treatment. Additional pairwise analyses demonstrated that Noggin completely inhibited all mRNAs whose expression was stimulated by BMP2, but had no effect on gene expression by BMP6 (Fig. 5A and Tables 2 and 3). In contrast, RGMc40 equivalently inhibited all transcripts induced by BMP2 or BMP6, but was generally less potent than Noggin in counteracting the actions of BMP2 (Fig. 5A and Tables 2 and 3). Of note, expression of ID1 and ID3 were reduced below basal levels by Noggin in the presence of BMP2, but were decreased substantially less than other transcripts by RGMc40. We cannot explain the enhanced potency of Noggin to block expression of ID1 or ID3, or the reduced efficacy of RGMc40. Except for these two outliers, the inhibitory potency of RGMc40 was generally about half of that of Noggin for genes whose expression was induced by BMP2 (Fig. 5A and Tables 2 and 3). To verify and extend the results of microarray studies, we tested the effects of Noggin and RGMc40-Fc on BMP-stimulated gene expression for ATOH8, SLC6A19, DUSPI, and GATA2 by semi-quantitative PCR. As also shown by microarray results, Noggin was more effective than RGMc40 in blocking the actions of BMP2, but was less effective in inhibiting BMP6-induced genes (Fig. 5B).
FIGURE 5.
RGMc40 inhibits BMP2- and BMP6-mediated gene expression. A, results of microarray analyses using RNA isolated from Hep3B cells treated with BMP2 or BMP6 (100 ng/ml) for 4 h. For the indicated samples growth factors were preincubated for 3 h at 20 °C with a 10-fold molar excess of Noggin-Fc or a 20-fold molar excess of RGMc40-Fc. Displayed is a heat map of genes identified as differentially expressed (>2-fold change versus un-treated; one-way analysis of variance, p < 0.02). The scale bar indicates the approximate fold-change versus un-treated. See supplemental Table S1 for additional gene information. B, analysis of ATOH8, SLC6A19, DUSPI, GATA2, and S17 mRNA levels by RT-PCR after incubation of cells with BMP2 or BMP6 for 4 h ± a 10-fold molar excess of Noggin-Fc or a 20-fold molar excess of RGMc40-Fc administered as in A.
TABLE 2.
Effects of Noggin and RGMc40 on genes induced by BMP2 in Hep3B cells (fold change)
| Gene | Gene name | BMP2 versus un-treated | Noggin versus BMP2 | RGMc versus BMP2 |
|---|---|---|---|---|
| ID1 | Inhibitor of DNA binding 1 | 13.2 | −29.4 | −2.0 |
| ID3 | Inhibitor of DNA binding 3 | 11.1 | −22.9 | −2.1 |
| SLC6A19 | Solute carrier family 6, member 19 | 7.2 | −6.4 | −2.9 |
| SMAD6 | SMAD family member 6 | 6.7 | −7.0 | −4.0 |
| SMAD7 | SMAD family member 7 | 4.3 | −4.6 | −2.1 |
| GHR | Growth hormone receptor | 4.1 | −3.8 | −1.4 |
| BAMBI | BMP and activin membrane-bound inhibitor homolog | 4.1 | −3.8 | −2.6 |
| urothelial cancer associated 1 | 4.1 | −4.2 | −2.2 | |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | 4.0 | −3.6 | −3.7 |
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | 3.9 | −4.3 | −3.0 |
| SLC29A3 | Solute carrier family 29, member 3 | 3.8 | −4.1 | −1.8 |
| DSE | Dermatan sulfate epimerase | 3.7 | −3.5 | −1.4 |
| SMAD9 | SMAD family member 9 | 3.7 | −3.9 | −2.1 |
| PFKFB3 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 3.6 | −3.6 | −2.1 |
| GPD1L | Glycerol-3-phosphate dehydrogenase 1-like | 3.5 | −3.3 | −1.8 |
| ATOH8 | Atonal homolog 8 | 3.4 | −4.1 | −2.4 |
| ID4 | Inhibitor of DNA binding 4 | 3.4 | −4.0 | −2.2 |
| GATA2 | GATA binding protein 2 | 3.2 | −3.7 | −2.4 |
| GATA3 | GATA binding protein 3 | 3.1 | −3.5 | −2.3 |
| SLCO2A1 | Solute carrier organic anion transporter family, memb. 2A1 | 3.1 | −2.7 | −1.8 |
TABLE 3.
Effects of Noggin and RGMc40 on genes induced by BMP6 in Hep3B cells (fold change)
| Gene | Gene name | BMP6 versus un-treated | Nogginaversus BMP6 | RGMc versus BMP6 |
|---|---|---|---|---|
| ID1 | Inhibitor of DNA binding 1 | 11.2 | −2.1 | |
| ID3 | Inhibitor of DNA binding 3 | 9.7 | −2.5 | |
| SLC6A19 | Solute carrier family 6, member 19 | 6.3 | −4.0 | |
| SMAD6 | SMAD family member 6 | 6.2 | −4.4 | |
| SMAD7 | SMAD family member 7 | 4.5 | −2.6 | |
| GHR | Growth hormone receptor | 4.0 | −1.7 | |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | 3.9 | −4.0 | |
| ATOH8 | Atonal homolog 8 | 3.9 | −3.5 | |
| BAMBI | BMP and activin membrane-bound inhibitor homolog | 3.8 | −3.0 | |
| DSE | Dermatan sulfate epimerase | 3.7 | −1.4 | |
| SLC29A3 | Solute carrier family 29, member 3 | 3.6 | −2.2 | |
| SMAD9 | SMAD family member 9 | 3.5 | −2.6 | |
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | 3.5 | −3.2 | |
| GPD1L | Glycerol-3-phosphate dehydrogenase 1-like | 3.4 | −2.1 | |
| urothelial cancer associated 1 | 3.3 | −2.4 | ||
| PFKFB3 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 3.3 | −2.4 | |
| GATA2 | GATA binding protein 2 | 3.1 | −2.4 | |
| GATA3 | GATA binding protein 3 | 3.1 | −2.5 |
a Noggin had no effect on any of these transcripts.
Selected Juvenile Hemochromatosis-associated RGMc Mutants Inhibit BMP2- and BMP6-mediated Gene Expression
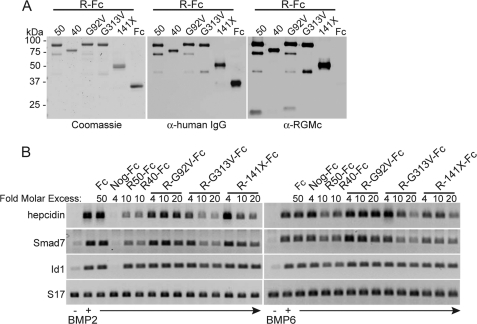
We next addressed the effects of selected disease-associated RGMc mutant proteins on BMP-activated signaling. We expressed and purified as IgG1-Fc fusion proteins the mouse versions of human RGMc amino acid substitution mutants G99V and G320V (RGMc-G92V-Fc and RGMc-G313V-Fc, respectively), as well as the truncation mutant 148X (mouse RGMc141X-Fc), as illustrated in Fig. 6A. Addition of a 10-fold molar excess of either RGMc50-Fc or RGMc40-Fc to Hep3B cells caused a ∼60% decrease in the extent of BMP-stimulated accumulation of transcripts encoding hepcidin and SMAD7, whereas a 50-fold molar excess of IgG1-Fc was ineffective, and a 4-fold excess of Noggin-Fc completely inhibited the actions of BMP2 but not BMP6 (Fig. 6B). RGMc-G92V-Fc did not inhibit the effects of either BMP, but surprisingly RGMc-G313V-Fc was as potent as wild-type RGMc50-Fc or RGMc40-Fc, particularly for BMP2, and RGMc141X-Fc was about half as effective on a molar basis (Fig. 6B). Because previous studies indicated that RGMc-G313V-Fc could bind BMP2 ∼10% as effectively as RGMc50-Fc, and that RGMc-G92V-Fc could not bind BMP2 (25), these results show that RGMc mutants inhibit BMP actions by direct growth factor binding, and indicate that there is a rough correlation between binding strength and inhibitory activity.
FIGURE 6.
Selected mutant RGMc proteins inhibit BMP-mediated gene expression. A, analysis of purified RGMc-Fc (R-Fc) fusion proteins or IgG1-Fc (Fc) after SDS-PAGE by Coomassie Blue staining (left panel), or by immunoblotting for IgG1-Fc (middle panel) or RGMc (right panel). RGMc proteins are as follows: 50, RGMc50; 40, RGMc40; G92V, RGMcG92V substitution mutant; G313V, RGMcG313V substitution mutant; 141X, RGMc141X truncation mutant. B, effects on gene expression: BMP2 or BMP6 (100 ng/ml) were preincubated for 3 h at 20 °C with 50-fold molar excess of IgG1-Fc, 4-fold molar excess of Noggin-Fc (Nog-Fc), 10-fold molar excess of R50-Fc or R40-Fc, or 4–20-fold molar excess of R-G92V-Fc, R-G313V-Fc, or R-141X-Fc, and then added to Hep3B cells for 4 h. Analysis of hepcidin, SMAD7, ID1, and S17 mRNA levels by RT-PCR is shown.
DISCUSSION
The discovery that mutations in the gene encoding HJV/RGMc resulted in the rapidly progressive iron overload disorder, JH (4), first implicated this protein in the regulation of whole body iron metabolism (5, 6). Further studies have shown that RGMc undergoes a series of biosynthetic and processing steps that result in four distinct mature protein species, including two cell-associated and two soluble isoforms (12, 14). The latter, here termed RGMc50 and RGMc40, have also been detected in the circulation (32). To date, the precise biological role of each RGMc species in iron metabolism has not been established. We now demonstrate that the two soluble RGMc isoforms are effective inhibitors of the actions of at least two classes of BMPs. We additionally show that the ability to interfere with BMP-mediated signaling extends to several JH-linked mutant RGMc proteins. These latter observations raise the hypothesis that disease severity in JH may vary depending on the type of mutation and the biochemical characteristics of the mutant protein, although no information is yet available regarding the biosynthesis, processing, or secretion of any mutant JH-linked HJV/RGMc protein in vivo.
BMPs are a diverse group within the transforming growth factor-β superfamily of growth factors (16, 17), and exert a wide range of biological effects on many cellular processes (16, 17). BMPs activate the signal transducers, Smads 1, 5, and 8, after binding to specific transmembrane receptors (16, 17), and Smads mediate many of the acute actions of BMPs on gene regulation (35). BMPs have been shown recently to promote the expression of the gene encoding the iron-regulatory hormone hepcidin in the liver (27, 28) and in cultured cells (15, 29, 31, 36), and it has been proposed that the BMP-Smad signaling pathway is a critical modulator of systemic iron metabolism via its effects on hepcidin production (27, 28, 37). We now find that soluble RGMc50 and RGMc40 can function as broad-based BMP inhibitors, and prevent both BMP2 and BMP6 from activating their receptors, as evidenced by the acute impairment of Smad phosphorylation, and the longer term reduction in gene activation that we observe. Our results thus extend previous studies showing that several different BMPs can bind to RGMc in vitro or in cell-based assays (15, 25, 27, 38), and other observations demonstrating that soluble RGMc40 can inhibit BMP2- and BMP4-mediated signaling and hepcidin gene expression in primary mouse hepatocytes and other cultured cells (31, 39).
Our ability to generate highly purified RGMc-IgG Fc fusion proteins has further allowed us to determine that RGMc50 and RGMc40 are fairly equivalent BMP inhibitors on a molar basis, and to demonstrate that their effects are similarly antagonistic to both BMP2 and BMP6, even though these two BMPs are only ∼61% identical in amino acid sequence (20), and preferentially activate different Type I and Type II receptors (21). Remarkably, in Hep3B cells both BMPs are nearly equivalently active, as measured by their ability to stimulate Smad phosphorylation and promote the expression of an identical cohort of ∼40 genes to the same extent in microarray profiling studies, indicating that several classes of receptors must be present in these cells.
The results obtained with RGMc50 or RGMc40 on BMP-mediated signaling and gene regulation contrast with the effects of Noggin, which in our hands was ∼5-fold more potent on a molar basis in inhibiting BMP2 than were either RGMc protein, but was ineffective toward BMP6. Among other previously described BMP antagonists, Chordin is relatively potent toward BMP2 and -4 (40) and follistatin is fairly effective against BMP7 (41), whereas DAN and the follistatin-related protein are fairly weak toward BMP2, -4, and -7 (42–44). Chordin-like and Sclerostin can block the actions of BMP6, but not BMP2 (45, 46), and CTGF and Nov family members primarily inhibit BMP2 and -4 (47, 48). Thus, soluble RGMc proteins appear to represent a fairly unique class of broad-spectrum BMP antagonists that target two different BMP subfamilies, and it will be of great interest once structural studies are completed to compare their mode of binding BMPs with those of other antagonists.
Several single-nucleotide mutations in the human HJV/RGMc gene that cause JH are predicted to encode RGMc proteins with single amino acid substitutions (4). In one of the more prevalent disease-associated mutants, glycine 320 is changed to valine (G320V (4), G313V in mouse RGMc (4)), and this protein can bind BMP2, although less effectively than wild type RGMc (25). We now find that purified recombinant mouse RGMc G313V can inhibit BMP2- and BMP6-stimulated hepcidin gene expression almost as effectively as wild type RGMc50. In contrast, the G92V mutation (G99V in human RGMc (4)), which cannot bind BMP2 (25), does not interfere with BMP-mediated signaling. Although a clinical relationship has not been established yet between a specific HJV genotype and disease phenotype in JH, and the precise levels of expression or secretion of mutated HJV/RGMc proteins in individuals with JH are unknown, it is conceivable that mutations that lead to the soluble RGMc species that bind BMPs may be more severe than those that do not, because of a greater inhibition of hepcidin production.
Very few studies to date have examined the actions of truncated forms of RGMc, such as those predicted from JH-associated HJV/RGMc gene mutations that cause frameshifts within the protein coding region (4). We now find that the mutant 141X truncation, consisting of ∼110 NH2-terminal residues of mature RGMc, when produced and purified as an IgG Fc fusion protein, retains the ability to inhibit the actions of both BMP2 and BMP6. These results allow us to hypothesize that a major domain for BMP binding is found within the NH2-terminal third of RGMc. Because the RGMc G92V mutant protein cannot not bind BMPs (25), and does not reduce their biological actions, we further postulate that a glycine or similarly sized aliphatic amino acid at this position is essential for interactions of RGMc with BMPs. Additional studies will be needed to define the critical determinants for BMP binding.
The two single-chain soluble RGMc species studied here are derived from the single-chain cell-associated 50-kDa isoform by two distinct proteolytic cleavages (32, 33). This cell-linked RGMc protein is attached to the membrane by a COOH-terminal glycosylphosphatidylinositol anchor, as is the two-chain RGMc species composed of ∼30 and ∼20 kDa subunits (12). It has been postulated that cell membrane-linked RGMc can function as a BMP co-receptor (15), although this hypothesis has not been tested rigorously, and it has not been established which of the two cell-associated RGMc isoforms is responsible for facilitating BMP actions, nor what the biochemical mechanisms might be. One of the challenges inherent in attempting to discern the biological effects of membrane-linked RGMc species is the presence of soluble isoforms, which in the case of BMP-mediated signaling will dampen any responses. Clearly new experimental strategies will be needed to help elucidate the distinct functions of each RGMc isoform in iron metabolism, and to define the biochemical basis for their biological effects.
In summary, through the use of highly purified recombinant fusion proteins, we have shown that soluble RGMc species are effective BMP antagonists, and appear to act by preventing access of at least two classes of BMPs to cell-surface receptors. Soluble RGMc proteins thus have a broader spectrum of inhibitory effects than Noggin, which primarily targets the BMP2/4 subclass (22), and other previously described BMP antagonists, which also have a limited range of activity (40–45). We additionally find that selected JH-associated RGMc mutant proteins also can inhibit BMP actions, raising the hypothesis that disease severity in JH may vary depending on potential functions of individual mutants. Moreover, deletion mapping studies with a JH-linked truncation mutant have allowed us to identify a minimal BMP binding domain within the NH2-terminal ∼110 amino acids of RGMc. Our results thus establish an experimental framework for discerning the biological roles of soluble RGMc proteins in BMP-mediated signaling, and for defining the critical amino acids and structural determinants responsible for binding these diverse BMPs.
Supplementary Material
Acknowledgments
Microarray assays were performed in the Affymetrix Microarray Core of the OHSU Gene Microarray Shared Resource. We thank Eric Olson and Hugh Arnold for training in GeneSifter® software.
This work was supported, in whole or in part, by National Institutes of Health Research Grants R01 DK042748 (to P. R.), F31 HL095271 (to M. N.), and WSF 0746589 (to U. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- JH
- juvenile hemochromatosis
- HJV
- hemojuvelin
- RGM
- repulsive guidance molecule
- BMP
- bone morphogenetic protein
- PNGaseF
- N-glycosidase F
- RT
- reverse transcription.
REFERENCES
- 1.Andrews N. C., Schmidt P. J. (2007) Annu. Rev. Physiol. 69, 69–85 [DOI] [PubMed] [Google Scholar]
- 2.Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004) Cell 117, 285–297 [DOI] [PubMed] [Google Scholar]
- 3.Beutler E. (2006) Annu. Rev. Med. 57, 331–347 [DOI] [PubMed] [Google Scholar]
- 4.Papanikolaou G., Samuels M. E., Ludwig E. H., MacDonald M. L., Franchini P. L., Dubé M. P., Andres L., MacFarlane J., Sakellaropoulos N., Politou M., Nemeth E., Thompson J., Risler J. K., Zaborowska C., Babakaiff R., Radomski C. C., Pape T. D., Davidas O., Christakis J., Brissot P., Lockitch G., Ganz T., Hayden M. R., Goldberg Y. P. (2004) Nat. Genet. 36, 77–82 [DOI] [PubMed] [Google Scholar]
- 5.Huang F. W., Pinkus J. L., Pinkus G. S., Fleming M. D., Andrews N. C. (2005) J. Clin. Invest. 115, 2187–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niederkofler V., Salie R., Arber S. (2005) J. Clin. Invest. 115, 2180–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuninger D., Kuzmickas R., Peng B., Pintar J. E., Rotwein P. (2004) Genomics 84, 876–889 [DOI] [PubMed] [Google Scholar]
- 8.Schmidtmer J., Engelkamp D. (2004) Gene Expr. Patterns 4, 105–110 [DOI] [PubMed] [Google Scholar]
- 9.Niederkofler V., Salie R., Sigrist M., Arber S. (2004) J. Neurosci. 24, 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samad T. A., Srinivasan A., Karchewski L. A., Jeong S. J., Campagna J. A., Ji R. R., Fabrizio D. A., Zhang Y., Lin H. Y., Bell E., Woolf C. J. (2004) J. Neurosci. 24, 2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babitt J. L., Zhang Y., Samad T. A., Xia Y., Tang J., Campagna J. A., Schneyer A. L., Woolf C. J., Lin H. Y. (2005) J. Biol. Chem. 280, 29820–29827 [DOI] [PubMed] [Google Scholar]
- 12.Kuninger D., Kuns-Hashimoto R., Kuzmickas R., Rotwein P. (2006) J. Cell Sci. 119, 3273–3283 [DOI] [PubMed] [Google Scholar]
- 13.Samad T. A., Rebbapragada A., Bell E., Zhang Y., Sidis Y., Jeong S. J., Campagna J. A., Perusini S., Fabrizio D. A., Schneyer A. L., Lin H. Y., Brivanlou A. H., Attisano L., Woolf C. J. (2005) J. Biol. Chem. 280, 14122–14129 [DOI] [PubMed] [Google Scholar]
- 14.Lin L., Goldberg Y. P., Ganz T. (2005) Blood 106, 2884–2889 [DOI] [PubMed] [Google Scholar]
- 15.Babitt J. L., Huang F. W., Wrighting D. M., Xia Y., Sidis Y., Samad T. A., Campagna J. A., Chung R. T., Schneyer A. L., Woolf C. J., Andrews N. C., Lin H. Y. (2006) Nat. Genet. 38, 531–539 [DOI] [PubMed] [Google Scholar]
- 16.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 17.Waite K. A., Eng C. (2003) Nat. Rev. Genet. 4, 763–773 [DOI] [PubMed] [Google Scholar]
- 18.Rosenzweig B. L., Imamura T., Okadome T., Cox G. N., Yamashita H., ten Dijke P., Heldin C. H., Miyazono K. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7632–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ten Dijke P., Yamashita H., Sampath T. K., Reddi A. H., Estevez M., Riddle D. L., Ichijo H., Heldin C. H., Miyazono K. (1994) J. Biol. Chem. 269, 16985–16988 [PubMed] [Google Scholar]
- 20.Kingsley D. M. (1994) Genes Dev. 8, 133–146 [DOI] [PubMed] [Google Scholar]
- 21.Lavery K., Swain P., Falb D., Alaoui-Ismaili M. H. (2008) J. Biol. Chem. 283, 20948–20958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman L. B., De Jesús-Escobar J. M., Harland R. M. (1996) Cell 86, 599–606 [DOI] [PubMed] [Google Scholar]
- 23.Keller S., Nickel J., Zhang J. L., Sebald W., Mueller T. D. (2004) Nat. Struct. Mol. Biol. 11, 481–488 [DOI] [PubMed] [Google Scholar]
- 24.Winkler D. G., Yu C., Geoghegan J. C., Ojala E. W., Skonier J. E., Shpektor D., Sutherland M. K., Latham J. A. (2004) J. Biol. Chem. 279, 36293–36298 [DOI] [PubMed] [Google Scholar]
- 25.Kuns-Hashimoto R., Kuninger D., Nili M., Rotwein P. (2008) Am. J. Physiol. Cell Physiol. 294, C994–C1003 [DOI] [PubMed] [Google Scholar]
- 26.Wilson E. M., Hsieh M. M., Rotwein P. (2003) J. Biol. Chem. 278, 41109–41113 [DOI] [PubMed] [Google Scholar]
- 27.Andriopoulos B., Jr., Corradini E., Xia Y., Faasse S. A., Chen S., Grgurevic L., Knutson M. D., Pietrangelo A., Vukicevic S., Lin H. Y., Babitt J. L. (2009) Nat. Genet. 41, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meynard D., Kautz L., Darnaud V., Canonne-Hergaux F., Coppin H., Roth M. P. (2009) Nat. Genet. 41, 478–481 [DOI] [PubMed] [Google Scholar]
- 29.Truksa J., Peng H., Lee P., Beutler E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10289–10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., Lin H. Y. (2007) J. Clin. Invest. 117, 1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin L., Valore E. V., Nemeth E., Goodnough J. B., Gabayan V., Ganz T. (2007) Blood 110, 2182–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuninger D., Kuns-Hashimoto R., Nili M., Rotwein P. (2008) BMC Biochem. 9, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L., Nemeth E., Goodnough J. B., Thapa D. R., Gabayan V., Ganz T. (2008) Blood Cells Mol. Dis. 40, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aschaffenburg R., Lewis M., Phillips D. C., Press E. M., Smith S. G., Sutton B. J., Mountford C. W. (1979) J. Mol. Biol. 135, 1033–1036 [DOI] [PubMed] [Google Scholar]
- 35.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 36.Zhang A. S., Yang F., Wang J., Tsukamoto H., Enns C. A. (2009) J. Biol. Chem. 284, 22580–22589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kautz L., Meynard D., Monnier A., Darnaud V., Bouvet R., Wang R. H., Deng C., Vaulont S., Mosser J., Coppin H., Roth M. P. (2008) Blood 112, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 38.Halbrooks P. J., Ding R., Wozney J. M., Bain G. (2007) J. Mol. Signal. 2, 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D. H., Zhou L. J., Zhou Z., Xie J. X., Jung J. U., Liu Y., Xi C. X., Mei L., Xiong W. C. (2010) Blood 115, 3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piccolo S., Agius E., Leyns L., Bhattacharyya S., Grunz H., Bouwmeester T., De Robertis E. M. (1999) Nature 397, 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iemura S., Yamamoto T. S., Takagi C., Uchiyama H., Natsume T., Shimasaki S., Sugino H., Ueno N. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dionne M. S., Skarnes W. C., Harland R. M. (2001) Mol. Cell. Biol. 21, 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu D. R., Economides A. N., Wang X., Eimon P. M., Harland R. M. (1998) Mol. Cell 1, 673–683 [DOI] [PubMed] [Google Scholar]
- 44.Tortoriello D. V., Sidis Y., Holtzman D. A., Holmes W. E., Schneyer A. L. (2001) Endocrinology 142, 3426–3434 [DOI] [PubMed] [Google Scholar]
- 45.Kusu N., Laurikkala J., Imanishi M., Usui H., Konishi M., Miyake A., Thesleff I., Itoh N. (2003) J. Biol. Chem. 278, 24113–24117 [DOI] [PubMed] [Google Scholar]
- 46.Nakayama N., Han C. E., Scully S., Nishinakamura R., He C., Zeni L., Yamane H., Chang D., Yu D., Yokota T., Wen D. (2001) Dev. Biol. 232, 372–387 [DOI] [PubMed] [Google Scholar]
- 47.Abreu J. G., Ketpura N. I., Reversade B., De Robertis E. M. (2002) Nat. Cell Biol. 4, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canalis E., Smerdel-Ramoya A., Durant D., Economides A. N., Beamer W. G., Zanotti S. (2010) Endocrinology 151, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.