FIGURE 1.
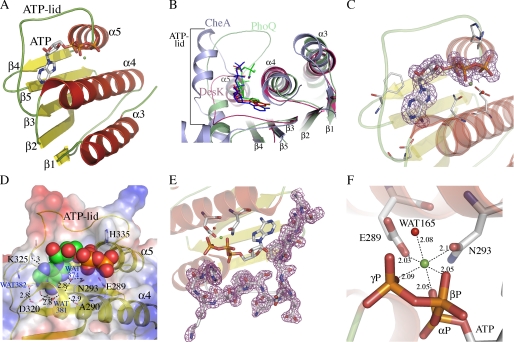
Structure of the ATP-binding domain of DesK and details of its nucleotide-binding pocket. A, schematic representation showing topology and secondary structure elements. α-Helices are colored red, β-strands are yellow, and loops are green. B, superposition of DesKABD (red tones), PhoQ (green tones), and CheA (blue tones) showing the exposure of the corresponding bound nucleotides to the solvent. Note the absence of secondary structure elements in the ATP lid of DesK. A surface representation of these differences is detailed in supplemental Fig. S2. C, 2mFobs − DFcalc electron density map contoured at 1σ, displayed around the ATP molecule, highlighting clear signal for the entire nucleotide. D, solvent-accessible surface representation of DesKABD, with the ATP moiety depicted as van der Waals balls. The surface is colored according to the mapping of electrostatic potential (red = negative, blue = positive). Key protein residues for ATP binding are highlighted in stick representation and explained in the text. The buried water H-bond network is also shown. Numbers correspond to distances in Å. E, 2mFobs − DFcalc map contoured at 1 σ, showing clear electron density for the entire ATP lid loop, ordered in the presence of ATP. F, close-up of the Mg2+ coordination site. Oxygens from the three ATP phosphates, residues Glu289 and Asn293, and water Wat165, are seen closing the octahedral coordination shell of the cation.