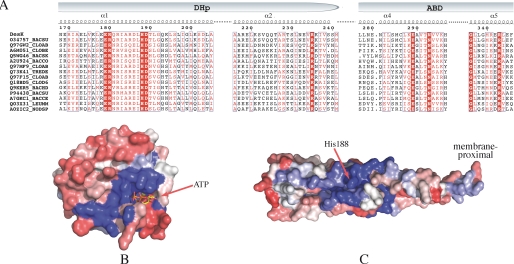
FIGURE 2.
Sequence alignments and conservation patterns of the HisKA_3 subfamily. A, selected region of a reduced multiple sequence alignment with sequences from the “HisKA_3” Pfam subfamily. Identical residues are shown in bold white on a red background; conserved residues are boxed in red. Pfam IDs are indicated on the left followed by a species code: BACSU, Bacillus subtilis; CLOAB, Clostridium acetobutylicum; CLOBE, Clostridium beijerinckii (strain ncimb 8052); BACSK, Bacillus clausii (ksm-k16); BACCO, Bacillus coagulans (36d1); TREDE, Treponema denticola; CLOD6, Clostridium difficile (630); BACHD, Bacillus halodurans; BACCE, Bacillus cereus cytotoxis (nvh 391–98); LEUMM, Leuconostoc mesenteroides mesenteroides (atcc 8293/ncdo 523); and NODSP, Nodularia spumigena (ccy 9414). B, sequence conservation/variability mapped onto the molecular surface of the ATP-binding domain of DesKC. Color coding is a range from red (variable) to blue (conserved) through an intermediate white. C, same as B, mapped onto the surface of the DHp domain of DesKC.