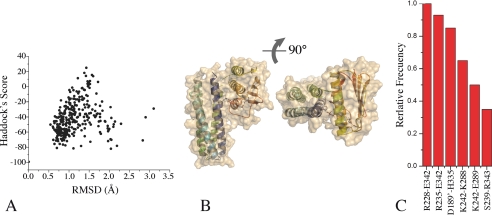
FIGURE 3.
Distance-restrained docking of DesKC ABD and DHp domains. A, the global Haddock score is plotted as a function of r.m.s.d. (calculated with respect to the best scoring model). Note that the best scoring models cluster at <1.5 Å r.m.s.d. B, three-dimensional structure of a representative high-scoring docked model of DesKC in the autophosphorylation state, in two orthogonal views. The solvent-accessible surface is rendered transparent to distinguish the relative organization of the domain-domain configuration (secondary structure elements are highlighted). Residues His188 and ATP are shown as sticks. C, relative frequency of interacting residue pairs in the best 10 docked structures. The histogram is limited to >30% frequencies. Engaged residues are listed, distinguishing with a single prime those belonging to the second protomer within the dimer.