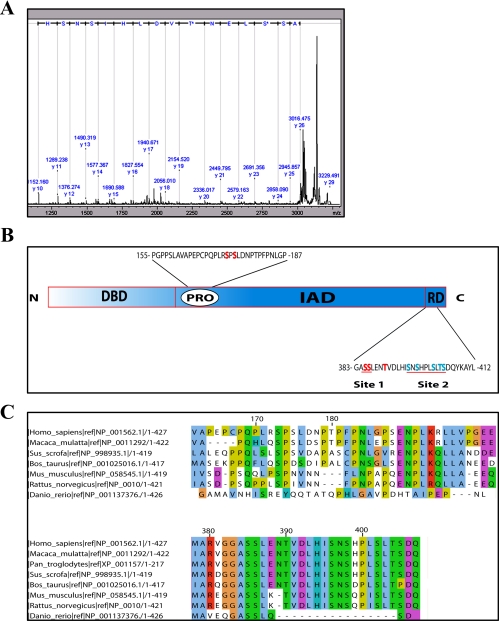
FIGURE 1.
Mass spectrometric identification of in vivo phosphorylation sites in IRF3. A, MS/MS spectrum of the doubly phosphorylated peptide 384–399. HEK293 cells were transfected with IRF3FLAG. 24 h later cells were infected with SV for 12 h. Lysates were immunoprecipitated with anti-FLAG-Sepharose and separated by BN-PAGE. Bands corresponding to IRF3 monomer or dimer were excised, and IRF3 was digested with trypsin prior to isolation of phosphopeptides using TiO2. The isolated peptides were analyzed by MALDI/TOF MS/MS. B, schematic overview of IRF3. The DNA-binding domain (DBD), proline-rich domain (PRO), IRF association domain (IAD), and regulatory domain (RD) are indicated. Residues in site 1 and site 2 are indicated (red line). C, alignment of IRF3 sequences from different species using JalView. The amino acids adjacent to Ser173 and Ser175 (upper panel) and amino acids in the C-terminal regulatory domain (lower panel) are shown.