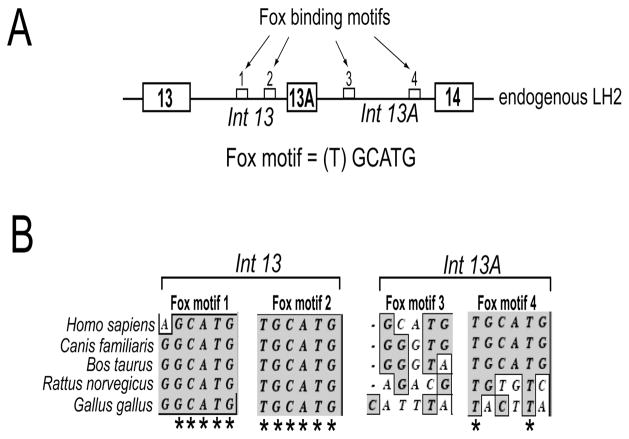
Figure 1. Clustal W sequence analysis of the flanking introns of exon 13A reveals that the Fox-2 binding motifs located in the upstream intron are highly conserved.
(A) Schematic (not drawn to scale) showing the genomic arrangement of the alternately spliced exon 13A and the constitutively spliced exons 13 and 14. The introns that flank exon 13A are also shown (Int 13 and Int 13A). The small white boxes represent the four Fox-2 binding motifs, shown by arrows, identified in the introns that flank exon 13A. The DNA sequence of the Fox binding motif is shown. The brackets around (T) indicate that it is the most variable base of the sequence. (B) Phylogenetic comparison of the sequence of the two Fox binding motifs present in intron 13 and two motifs present in intron 13A from five different species (humans, dog, cow, rat and chicken). Only the sequences of the two Fox binding motifs present in intron 13 and two motifs present in intron 13A are shown. The asterisks (*) denote complete sequence conservation across the five species.