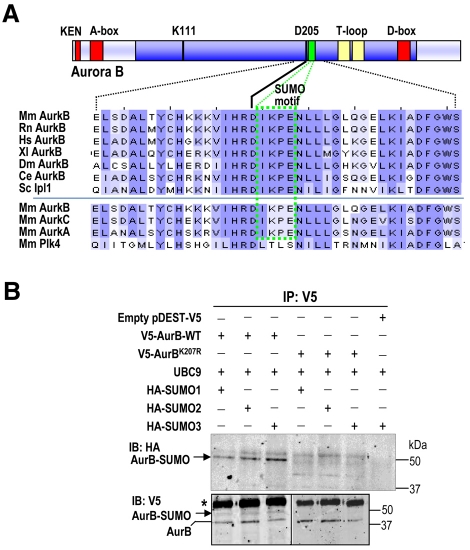
Fig. 1.
A SUMOylation motif in Aurora B. (A) Multiple sequence alignment of Aurora B proteins from different species using Clustal software. Mm, Mus musculus; Rn, Rattus norvegicus; Hs, Homo sapiens; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, single Aurora family member (Ipl1) from Saccharomyces cerevisiae. The SUMO motif (green box) is highly conserved in all these Aurora-B proteins, as well as in Aurora A and Aurora C, but it is not present in Polo-like kinase 4 (Plk4), a close mitotic relative of these mitotic kinases. The kinase domain is shown in dark blue, whereas the degradation motifs (KEN, A- and D-boxes) are represented as red boxes. Note that the SUMO motif is located one residue downstream of the aspartate (D205) that is crucial for ATP binding. Residue numbers correspond to the mouse Aurora-B protein. (B) In vivo SUMOylation of Aurora B at K207. HEK293 asynchronous cells were transiently transfected with vectors expressing V5-Aurora-B WT, Aurora BK207R, HA-SUMO1, -SUMO2 or -SUMO3 and the E2 SUMO ligase UBC9, as indicated. The empty vector pDEST-V5 was used as a negative control. V5-Aurora-B proteins were immunoprecipitated (IP) 48 hours after transfection using anti-V5 antibodies and immunoblotted (IB) with antibodies against the HA and V5 tags. Aurora-B-SUMO conjugates are found after detection of the HA and V5 tags in cells transfected with WT V5-Aurora-B but not in cells transfected with the V5-Aurora-BK207R mutant form. V5-Aurora-B immunoprecipitated in all lanes. Mouse IgGs are indicated with an asterisk.