Fig. 2.
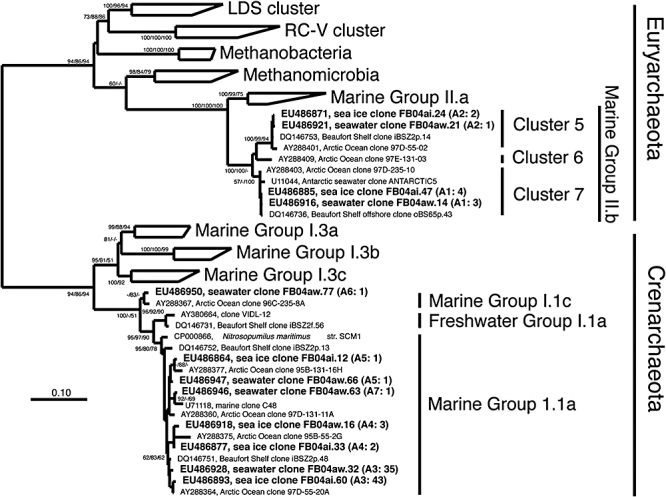
Phylogenetic tree of archaeal 16S rRNA gene sequences. Tree topology was defined by the consensus of 1000 maximum parsimony bootstrap replications utilizing 511 parsimony-informative nucleotides. Branch lengths were defined by Tamura-Nei distances calculated from 807 hypervariable-masked nucleotides. Node values indicate percentage of 10 000 distance, 1000 maximum parsimony and 100 maximum likelihood bootstrap replications respectively; only bootstrap values greater than 50% are shown. One sequence from each phylotype (defined by > 99% similarity) from each library in this study is shown in bold, followed in parentheses by the phylotype and number of sequences from each library within that phylotype.
