Abstract
The therapeutic efficacy of sulfadoxine-pyrimethamine (SP) in treating uncomplicated Plasmodium falciparum malaria is unevenly distributed in Colombia. The Andes mountain range separates regions in the west where malaria is endemic from those in the east and constitutes a barrier against gene flow and the dispersal of parasite populations. The distribution of dhfr and dhps genotypes of 146 P. falciparum samples from the eastern Amazon and Orinoco basins and Northwest and Southwest Pacific regions of Colombia was consistent with the documented levels of therapeutic efficacy of SP. The diversity of four dhfr- and dhps-linked microsatellites indicated that double- and triple-mutant alleles for both resistance loci have a single origin. Likewise, multilocus association genotypes, including two unlinked microsatellite loci, suggested that genetic exchanges between the eastern Orinoco and Northwest Pacific populations has taken place across the Andes, most probably via migration of infected people.
Enzymes involved in folate metabolism are targeted by the anifolate antimalarial drugs. Pyrimethamine targets the enzyme dihydrofolate reductase (DHFR) and, in combination with sulfadoxine, which targets the enzyme dihydropteroate synthase (DHPS), has been widely used as first-line treatment for uncomplicated Plasmodium falciparum malaria worldwide. In South America, pyrimethamine was introduced and used as a mass treatment in the 1950s in Venezuela (10) until drug-resistant cases were detected (16). By 1968, pyrimethamine-resistant/sulfadiazine-sensitive parasites were documented in Brazil, Venezuela, and Colombia (28). The sulfadoxine-pyrimethamine (SP) combination was introduced in parts of South America in the 1970s and was used until 1981 in Colombia as an alternative to chloroquine, to which resistance was widespread (8). Soon after its introduction, treatment failure was reported, and resistance rapidly disseminated in the Amazon and Orinoco basins (12). In Colombia, SP resistance is unevenly distributed. High SP resistance levels (80%) have been consistently reported from the Colombian Amazon basin (21), while moderate levels (6% to 24%) are observed in the Caribbean, the Cauca Valley, and northwestern regions (3, 4). This contrasts with regions on the southern Pacific coast where the SP combination is still efficacious (13).
P. falciparum resistance to pyrimethamine is acquired by the progressive accumulation of mutations at the dhfr locus. The S108N substitution is initially required for the acquisition of the resistant phenotype; in this genetic context, resistance increases with the accumulation of additional mutations at position N51I or C59R, and still higher levels of resistance are reached with the further acquisition of mutations at positions C50R and I164L (7, 14, 15). Resistance to sulfadoxine also depends on the progressive accumulation of mutations in the dhps locus at codons 436, 437, 540, 581, and 613. The A437G mutation is initially required and is followed by mutations at codons A581G, S436A, K540E, and A613S, which confer incrementally higher levels of resistance (14).
The current pattern of P. falciparum SP resistance in Colombia raises questions as to the nature, frequency, and origin of the circulating dhfr and dhps genotypes. Analysis of microsatellite markers linked to dhfr and dhps has shown that some mutant resistance alleles in Asia and Africa have become globally dispersed (27), while unlinked polymorphic loci on different chromosomes have suggested that the clonal expansion of a few resistant parasite genotypes underlies the spread of drug resistance in South America (6). Interestingly, in Colombia, the Andes mountain range separates the Pacific coast in the west from the Amazon and Orinoco basins in the east and may constitute a barrier against genetic exchanges. Here, we analyze the genetic nature and origin of P. falciparum SP resistance in Colombia. The results will help to the design strategies to prevent the spread of drug resistance in Colombia and eventually other Andean countries.
MATERIALS AND METHODS
P. falciparum samples.
Blood samples from subjects with thick-smear-confirmed uncomplicated P. falciparum malaria who took part in epidemiological studies carried out by Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM) and Instituto Nacional de Salud (INS) during the years 1999 to 2007 were stored on filter paper. A total of 146 samples representing four geographical regions of Colombia were selected: (i) Northwestern Pacific coast, Quibdó, Department of Chocó (n = 30; lat 5.694722, long −76.661111); (ii) Southwestern Pacific coast, Tumaco, Department of Nariño (n = 30; lat 1.798611, long −78.815556); (iii) Amazon basin, Department of Amazonas (n = 43, distributed as n = 14 from Leticia, lat −4.215278, long −69.940556; n = 16 from Tarapacá, lat −2.878611, long −69.744167; n = 3 from La Pedrera, lat −1.3002778, long −69.5638889; and n = 10 from an unknown locality in Amazonas Department); and (iv) Orinoco basin, Departments of Meta and Guaviare (n = 31 from Meta, lat 3.01236, long −73.154; and n = 12 from Guaviare, lat 2.570833, long −72.640278) (see Table S1 in the supplemental material). This study was approved by the Ethical Review Committee of CIDEIM.
DNA extraction, dhfr and dhps genotyping, sequencing, and microsatellite analysis.
Genomic DNA for PCR-restriction fragment length polymorphism (RFLP) analysis from P. falciparum samples stored in filter paper was extracted and sequenced as described previously (29). Polymorphic positions in the dhfr (positions 51, 59, 108, and164) and dhps (positions 437, 540, and 581) genes were genotyped as described previously (25; http://medschool.umaryland.edu/CVD/2002_pcr_asra.asp). To confirm the restriction enzyme analysis, 11 samples were sequenced for dhfr, 8 for dhps, and 10 for both genes after nested PCRs as described previously (23, 26). Genotyping of 6 microsatellite loci was performed in all samples: 2 dhfr-linked microsatellite loci (mDHFR and MA1, located 0.3 kb and 5.3 kb upstream, respectively, from codon 108 of the dhfr gene in chromosome 4 [chr4]), 2 dhps-linked microsatellites (m0.8 and m4.3, located 0.8 and 4.3 kb downstream, respectively, of the gene in chr8) (26), and 2 unlinked microsatellites (PfPK2 and Polyα), located in chr12 and chr4, respectively (1). The microsatellites were amplified using a seminested PCR, and the products were analyzed as previously described (26). When more than one allele was present at a locus, the allele with the higher peak was scored if the minority peak was less than 50% of the majority peak. If the minority peak exceeded the 50% cutoff value, the sample was not taken into account for the locus. Twenty-two samples out of 147 showed mixed infections, 14 of which were discarded for a given locus.
Resistance alleles, resistance genotypes, allelic haplotypes, and multilocus associations.
Combinations of dhfr and dhps resistance alleles were defined as the resistance genotype. Allelic haplotypes comprised dhfr and its linked microsatellite alleles (at mDHFR and MA1 microsatellite loci) or dhps and its linked microsatellite alleles (at m0.8 and m4.3 microsatellite loci). When linked microsatellites were combined with unlinked microsatellites, this was defined as the multilocus association genotype. Different multilocus association genotypes representing various degrees of linkage were taken into account as follows: (i) complete multilocus association genotype (i.e., allelic linkage at all six microsatellite loci) and (ii) partial multilocus associations (i.e., allelic linkage in two of the three above-mentioned pairs of microsatellite loci). Haplotypes and multilocus association genotypes were coded with numbers, and they are listed in full in Table S1 in the supplemental material.
Statistical analysis, linkage disequilibrium, and population differentiation.
Allelic, haplotypic, and multilocus association genotype diversity was calculated as Nei's unbiased gene diversity estimate (19) as implemented in the Microsatellite toolkit (22). Statistical analysis of population differentiation was performed using Arlequin 3.1 software (9). FST indexes (Wright's FST; fixation index) were calculated for neutrally evolving, nonlinked microsatellite loci (PfPK2 and Polyα) as pairwise comparisons between the four populations.
Allele frequency maps.
Resistance allele frequencies were calculated using the Microsatellite toolkit (22) and were used as input for maps (the frequencies are presented in Table S2 in the supplemental material). Samples missing an allele, because of either failed sequencing reactions or restriction enzyme digestion, were not included in the maps. Frequencies were arranged into those originating before 2006 and those originating in 2006 or after. Specific localities and dates of sampling are presented in Table S1 in the supplemental material.
RESULTS
dhfr and dhps circulating genotypes.
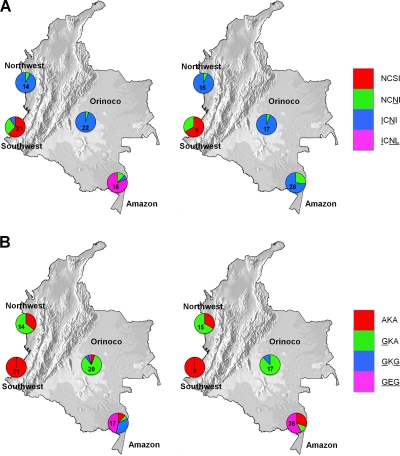
Of the 146 samples processed, 135 yielded interpretable results for both genes. There were 5 failures in the dhfr locus only and 2 in the dhps locus only. Figure 1 shows that in southwest and northwest Colombia the circulating dhfr alleles are wild-type NCSI (51/59/108/164) single NCNI (mutations are underlined) and double ICNI mutants, while in eastern Colombia (in the Amazon and Orinoco basins), single NCNI, double ICNI, and triple ICNL mutant alleles were found. Likewise, at dhps there were regional differences between eastern and western sites. Western sites were characterized by a mixture of sensitive AKA (437/540/581) and single GKA mutants, while eastern populations had double GKG and triple GEG mutants, as well. In the Southwestern Pacific coast, where SP treatment is still efficacious, there is a high prevalence of the wild-type genotypes dhfr NCSI (19/30; 63.3%) and dhps AKA (100%). In the Northwest Pacific coast, the majority of circulating parasites were dhfr ICNI double mutants (27/29; 93.1%) and wild type (AKA) (10/19; 52.6%) and single GKA dhps mutants (19/29; 65.5%). In the Orinoco basin, the majority of genotypes were dhfr ICNI double (37/39; 94.8%) and single dhps GKA (31/36; 86.1%) mutants. Only a single sample (from Vistahermosa, Meta, 2005) displayed the triple dhps GEG genotype. The dhfr double ICNI/dhps single GKA allele was the most common in both the northwest (18/29; 62.1%) and Orinoco (29/35; 83%). In the Amazon, a high proportion (13/16; 81.2%) of dhfr ICNL triple mutants was observed before 2006, but they disappeared in 2006 or after as a result of an increase of NCNI single (2/16, 12.5%, to 7/26, 26.9%) and ICNI (from 1/16, 6.3%, before to 19/26, 73.1%, in 2006 or after) double mutants. Similarly, dhps double GKG mutants disappeared in 2006 and after (6/17; 35.3%, before 2006), while triple dhps GEG mutants increased slightly (8/17, 47%, before 2006 to 15/26, 57.7%, in 2006 or after). Sequencing of eight dhfr ICNL triple-mutant samples showed that all bear the “Bolivian repeat” (BR) (a 15-bp repeat between codons 30 and 31 with no apparent effect on drug resistance). In addition, two sequenced samples from Amazonas had a mutation at codon 50 in the dhfr gene, and in both cases, it was found in the RICNI (50R/51I/59C/108N/164I) triple-mutant allele.
FIG. 1.
Allele frequencies for dhfr (positions 51, 59, 108, and 164) and dhps (positions 437, 540, and 581) genes in the eastern Amazonas and Orinoco basins and Northwest and Southwest Pacific, Colombia. dhfr resistant alleles: wild type (NCSI), single mutant (NCNI), double mutant (ICNI), and triple mutant (ICNL). dhps resistant alleles: wild type (AKA), single mutant (GKA), double mutant (GKG), and triple mutant (GEG). (A) dhfr allele frequencies. (B) dhps allele frequencies.(Left) Allele frequencies between 1999 and 2005. (Right) Allele frequencies between 2006 and 2007.
Microsatellite diversity and origins of resistance genotypes.
Twelve dhfr-linked microsatellite haplotypes (coded 1 to 12) were identified (see Table S1 in the supplemental material). dhfr ICNI double mutants were found in all four regions associated with 5 different dhfr-linked microsatellite haplotypes (“2,” “4,” “5,” “11,” and “12”); haplotype “4” was highly prevalent in all four regions (from 38,5% in the southwest to 95,7% in the northwest). Except for haplotype “12,” which can be explained as the result of a recombination event with other circulating parasites, the haplotypes were represented by a single sample and were related to the prevalent haplotype either by their close allele sizes or by sharing one of the two alleles (Table 1). This suggests that they all have a common origin. dhfr ICNL triple mutants were associated with three different dhfr-linked microsatellite haplotypes (“1,” “2,” and “7,”) two of which (“1” and “7”) were found in a single sample. The prevalent haplotype (“2”) shares a common allele with haplotype “1” at the MA1 locus; the two alleles at the DHFR locus are closely related in size (Table 1). Futhermore, sequencing of a random sample of 7 individuals, including haplotypes “7” and “8” and representing three different resistance genotypes, showed that they all contain the “Bolivian repeat,” indicating that the majority of dhfr triple mutants have a single origin.
TABLE 1.
dhfr-linked microsatellite haplotypes at double- and triple-mutant dhfr alleles
| Origin | No. of samples | dhfr allelec | mDHFRd (0.3 kb) | MA1e (5.3 kb) | Haplotype codef |
|---|---|---|---|---|---|
| SWPa | 2 | ICNI | 98 | 201 | 4 |
| NWPb | 20 | ICNI | 98 | 201 | 4 |
| 1 | ICNI | 96 | 201 | 2 | |
| Orinoco | 21 | ICNI | 98 | 201 | 4 |
| 1 | ICNI | 104 | 201 | 11 | |
| 1 | ICNI | 98 | 203 | 5 | |
| Amazonas | 1 | ICNI | 125 | 221 | 12 |
| 18 | ICNI | 98 | 201 | 4 | |
| Amazonas | 18 | ICNL | 102 | 221 | 8 |
| 1 | ICNL | 100 | 221 | 7 | |
| 1 | ICNL | 96 | 199 | 1 |
SWP, Southwest Pacific.
NWP, Northwest Pacific.
Double (ICNI) and triple (ICNL) mutant alleles at positions 50, 51, 108, and 164 of the dhfr locus.
Sizes (bp) of PCR allelic products at the mDHFR microsatellite locus. In parentheses, the distance upstream from the 108 codon is indicated.
Sizes (bp) of PCR allelic products at the MA1 microsatellite locus. In parentheses, the distance upstream from the 108 codon is indicated.
Each microatellite haplotype was coded as in Table S1 in the supplemental material.
Eighteen dhps-linked microsatellites haplotypes (coded 13 to 30) were identified (see Table S1 in the supplemental material). dhps GKG double mutants were associated with 2 dhps-linked microsatellite haplotypes (“17” and “19”) and GEG triple mutants with only one (“19”), indicating a single origin for the dhps GKG double and GEG triple mutants (Table 2). Single dhfr NCNI and dhps GKA mutants are related to 3 and 12 different linked microsatellite haplotypes, respectively. The large haplotypic diversity of dhfr- and dhps-linked microsatellites (average Hz, 0.813; n = 42) and the presence of eastern- and western-specific haplotypes suggest that single-mutant genotypes for both resistance loci have arisen on several occasions.
TABLE 2.
dhps-linked microsatellite haplotypes at double- and triple-mutant dhps alleles
| Origin | No. of samples | dhps allelea | m0.8b (−0.8 kb) | m4.3c (−4.3 kb) | Haplotype coded |
|---|---|---|---|---|---|
| Orinoco | 3 | GKG | 115 | 104 | 19 |
| Amazonas | 1 | GKG | 113 | 102 | 17 |
| 3 | GKG | 115 | 104 | 19 | |
| 23 | GEG | 115 | 104 | 19 |
Double (GKG) and triple (GEG) mutant alleles at positions 437, 540, and 581 of the dhps locus.
Sizes (bp) of PCR products at m0.8 microsatellite locus. In parentheses, the distance downstream from the 437 codon is indicated.
Sizes (bp) of PCR products at the m4.3 microsatellite locus. In parentheses, the distance downstream from the 437 codon is indicated.
Each microsatellite haplotype was coded as in Table S1 in the supplemental material.
Genetic differentiation and migration.
The largest number of shared multilocus association combinations occurred across the Andes, between northwestern and eastern Orinoco populations (see Table S3 in the supplemental material). Three complete multilocus associations (i.e., dhfr- and dhps-linked microsatellite haplotypes and the Polyα and PfPK2 unlinked microsatellite combination) were shared between the northwest and Orinoco, two between Amazonas and Orinoco, and one between the northwest and southwest populations. If only the combination of neutral loci (Polyα and PfPK2) is taken into account, four combinations are observed to be shared between the northwest and Orinoco, two between Amazonas and Orinoco, and two between the northwest and southwest populations.
It is expected that the introduction of new alleles into a population increases the genetic diversity and diminishes the differentiation between populations. The highest diversity values (expected heterozygosities) for the unlinked-microsatellite combination and the complete multilocus association genotype were consistently observed in the northwestern population (Table 3). Diversity values for all individual loci are presented in Table S4 in the supplemental material. Similarly, pairwise differentiation indices showed evidence of significant population structure. The Orinoco/northwest and the northwest/southwest populations were the less differentiated populations (FST values:, 0.094 and 0.070, respectively). In contrast, the most differentiated populations were the Amazon and Orinoco subpopulations (FST, 0.252) and the Tumaco population (average FST value, 0.235) relative to the eastern subpopulations (Table 4).
TABLE 3.
Diversity values (expected heterozygosities) for the unlinked microsatellite combination and the complete multilocus association genotypea
| Populationb | Unlinked-microsatellite association genotypec | Complete multilocus association genotyped |
|---|---|---|
| Amazonas | 0.691 (n = 38) | 0.760 (n = 33) |
| Orinoco | 0.640 (n = 25) | 0.860 (n = 19) |
| NWP | 0.915 (n = 27) | 0.971 (n = 21) |
| SWP | 0.837 (n = 29) | 0.909 (n = 24) |
Diversity values for all individual loci are presented in Table S3 in the supplemental material.
NWP, Northwest Pacific; SWP, Southwest Pacific.
PfPK2 and Polyα.
mDHFR, MA1, m0.8, m4.3, PfPK2,and Polyα.
TABLE 4.
Pairwise fixation index (FST) values between and within eastern (Amazonas and Orinoco) and western (Southwest and Northwest Pacific) Colombian P. falciparum populations based on the unlinked-microsatellite (PfPK2 and Polyα) multilocus association genotype
| Population paira | FST (mean PfPK2 and Polyα loci)b |
|---|---|
| Amazonas/Orinoco | 0.252 |
| Amazonas/NWP | 0.190 |
| Amazonas/SWP | 0.236 |
| Orinoco/NWP | 0.094 |
| Orinoco/SWP | 0.234 |
| NWP/SWP | 0.070 |
NWP, Northwest Pacific; SWP, Southwest Pacific.
FST values are significant at the 5% level.
DISCUSSION
The distribution of dhfr and dhps genotypes found in this study is consistent with the observed geographical differences in the therapeutic efficacies of SP against uncomplicated P. falciparum malaria in Colombia. Where SP is still efficacious (southwestern Colombia), the majority of circulating genotypes were wild type for both dhfr and dhps genes; where there is moderate resistance to SP (northwestern Colombia), the majority of circulating genotypes were dhfr double (ICNI) and dhps single (GKA) mutants. In the Amazon basin, where 80% SP treatment failures have been documented (21), quadruple-, quintuple-, and sextuple-mutant resistance genotypes were observed (Fig. 1). The absence in the Amazon of the highly resistant dhfr ICNL genotypes and the presence of the dhps wild type (AKA) after 2006 suggests the relaxation of drug pressure in the area. However, SP was removed from the national antimalarial drug policy only at the end of 2007. Accordingly, it is important to consider, due to the high human mobility in the area, the impact of Peru's abandonment of SP to treat P. falciparum malaria in their Amazon region in 2001 (20).
A number of genotypes associated with SP resistance have been described from South America (see reference 18 for a review). Our results concur with previous observations from South American samples suggesting a common origin for the so-called secondary antifolate resistance mutants. Samples bearing the dhfr ICNL triple-mutant allele from Amazonian Peru and Bolivia have the same or very similar dhfr-linked microsatellite allele sizes, as well as the Bolivian repeat, suggesting a common origin (2, 6, 30). Contrary to observations in other places in South America (2, 30), our data also suggest a single origin for Colombian dhfr ICNI double mutants. The triple mutant RICNI, with an apparently different origin and conferring midlevel resistance, has been reported from Brazil, Bolivia, Peru, Suriname, and Venezuela (2, 5, 24).
Likewise, as previously observed, dhps double GKG and triple GEG mutants seem to have a single origin in the Amazon and Orinoco basins (2, 24, 30) and have not disseminated into western Colombia. The single dhps GKA mutant has been reported in Brazil, Colombia, Peru, Suriname, and Venezuela; the double mutant GKG from Brazil, Peru, and Venezuela; and the triple mutant GEG in Amazonian Brazil, Bolivia, Peru, Suriname, and Venezuela. Analyses of microsatellites closely linked to the resistance genes have shown that double and triple mutants seem to have a common origin (2, 6, 11, 17, 30).
The number of shared haplotypes and multilocus association genotypes and the high levels of genetic diversity values for the Northwest Pacific (0.915) population, as well as the low FST values (0.094) between the Northwest Pacific and Orinoco populations, indicate the occurrence of genetic exchanges across the Andes mountain range between specific eastern (Orinoco) and western (Quibdo) localities in Colombia, most likely due to human migration. There is also evidence for genetic exchanges within each of the eastern and western populations. Since it is expected that the probability of a particular genotype invading another population increases with its frequency, genotypes with the highest frequency values are thought to have the highest probability of invading. This, as well as the time of emergence of a particular haplotype or multilocus association genotype, should give some indication as to the direction of migration. Following these criteria, our data indicate that migration has taken place in both directions (east and west of the Andes). Relatively high-frequency haplotypes in the Northwest Pacific population before 2006 are observed at relatively high frequencies in Orinoco and the Southwest Pacific after 2006, suggesting a flow of migration from the Northwest Pacific to the Southwest Pacific and Orinoco. Similarly, putatively clonal, complete multilocus association genotypes observed at a high frequency (60%) in Orinoco before 2006 are observed in the Northwest after 2006 (a detailed account of the dates of collection for each of the samples used in this study can be found in Table S1 in the supplemental material). The highest diversity values for the multilocus association genotypes occur in the Northwest Pacific population (Table 3). This suggests that although migration occurs in both directions, the flow may be higher from Orinoco to the Northwest Pacific.
In conclusion, these findings support the hypothesis of the expansion of P. falciparum SP-resistant populations and highlight its importance for the formulation of multiregional antimalarial drug policies to deter the spread of drug resistance. Mapping human migration routes and understanding human migration patterns east and west of the Andes would be useful to decide whether malaria control strategies targeted to internal migrants are feasible. Further studies are required to determine the consequences of SP withdrawal in each of the regions and the magnitude and directions of genetic flow between populations on a larger scale (e.g., South America).
Supplementary Material
Acknowledgments
This work was supported by COLCIENCIAS contract number 366-2006 and WHO/TDR contract number T16/181/658 ID no. A41403.
We thank the managers of the malaria control programs in Amazon, Chocó, Nariño, Meta, and Guaviare for their collaboration in the sample collection, as well as Santiago Nicholls for support with the acquisition of samples.
Footnotes
Published ahead of print on 24 May 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Anderson, T. J., X. Z. Su, M. Bockarie, M. Lagog, and K. P. Day. 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119:113-125. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, D. J., A. M. McCollum, S. M. Griffing, C. Salas, V. Soberon, M. Santolalla, R. Haley, P. Tsukayama, C. Lucas, A. A. Escalante, and V. Udhayakumar. 2009. Dynamics of malaria drug resistance patterns in the Amazon basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob. Agents Chemother. 53:2042-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, S., J. Carmona-Fonseca, J. G. Pineros, A. Rios, T. Alvarez, G. Alvarez, and A. Tobon. 2006. Therapeutic efficacy test in malaria falciparum in Antioquia, Colombia. Malar. J. 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair, S., L. L. Lacharme, J. C. Fonseca, and A. Tobon. 2001. Resistance of Plasmodium falciparum to 3 antimalarials in Turbo (Antioquia, Colombia), 1998. Rev. Panam. Salud Publica 9:23-29. [DOI] [PubMed] [Google Scholar]
- 5.Contreras, C. E., J. F. Cortese, A. Caraballo, and C. V. Plowe. 2002. Genetics of drug-resistant Plasmodium falciparum malaria in the Venezuelan state of Bolivar. Am. J. Trop. Med. Hyg. 67:400-405. [DOI] [PubMed] [Google Scholar]
- 6.Cortese, J. F., A. Caraballo, C. E. Contreras, and C. V. Plowe. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999-1006. [DOI] [PubMed] [Google Scholar]
- 7.Cortese, J. F., and C. V. Plowe. 1998. Antifolate resistance due to new and known Plasmodium falciparum dihydrofolate reductase mutations expressed in yeast. Mol. Biochem. Parasitol. 94:205-214. [DOI] [PubMed] [Google Scholar]
- 8.Espinal, T. C. A., C. G. T. Cortes, P. Guerra, and A. E. Arias. 1985. Sensitivity of Plasmodium falciparum to antimalarial drugs in Colombia. Am. J. Trop. Med. Hyg. 34:675-680. [DOI] [PubMed] [Google Scholar]
- 9.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 10.Gabaldon, A., L. Guerrero, C. Balestrini, B. G. Martin, R. Grossi, S. Maberti, T. Mendicoa, C. Psinakis, E. Russian, V. Salterini, M. Tabachiera, and A. Irigoyen. 1959. An attempt to eradicate malaria by the weekly administration of pyrimethamine in areas of out-of-doors transmission in Venezuela. Am. J. Trop. Med. Hyg. 8:433-439. [DOI] [PubMed] [Google Scholar]
- 11.Galindo, J. A., C. F. A. Knudson, R. S. Nicholls, and A. P. Guerra. 2010. Mutaciones puntuales en los genes dhfr y dhps de Plasmodium falciparum de tres regiones endémicas de malaria en Colombia. Biomédica 30:56-64. [PubMed] [Google Scholar]
- 12.Godoy, G. A., G. S. Volcan, R. Guevara, C. Medrano, J. Castro, and A. Texeira. 1977. Venezuelan strains of Plasmodium falciparum resistant to sulfa and pyrimethamine as demonstrated by in vitro test. Rev. Latinoam. Microbiol. 19:229-231. [PubMed] [Google Scholar]
- 13.González, I. J., J. O. Padilla, L. E. Giraldo, and N. G. Saravia. 2003. Efficacy of amodiaquine and sulfadoxine/pyrimethamine in the treatment of malaria not complicated by Plasmodium falciparum in Narino, Colombia, 1999-2002. Biomedica 23:38-46. [PubMed] [Google Scholar]
- 14.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 15.Hyde, J. E., S. Dittrich, P. Wang, P. F. Sims, V. de Crecy-Lagard, and A. D. Hanson. 2008. Plasmodium falciparum: a paradigm for alternative folate biosynthesis in diverse microorganisms? Trends Parasitol. 24:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maberti, S. 1960. The development of resistance to pyrimethamine. Presentation of 15 cases studied in Trujillo, Venezuela. Arch. Venez. Med. Trop. Parasitol. Med. 3:239-259. [PubMed] [Google Scholar]
- 17.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mita, T., K. Tanabe, and K. Kita. 2009. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol. Int. 58:201-209. [DOI] [PubMed] [Google Scholar]
- 19.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY.
- 20.Neyra, D., C. Cabezas, and T. K. Ruebush II. 2003. El proceso de adecuación y cambio en la política del tratamiento de la Malaria por Plasmodium falciparum en el Perú, 1990-2001. Rev. Peru Med. Exp. Salud Pública 20:162-171. [Google Scholar]
- 21.Osorio, L., L. D. P. Perez, and I. J. Gonzalez. 2007. Assessment of the efficacy of antimalarial drugs in Tarapaca, in the Colombian Amazon basin. Biomedica 27:133-140. [PubMed] [Google Scholar]
- 22.Park, S. D. E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis, University of Dublin, Dublin, Ireland.
- 23.Pearce, R. J., H. Pota, M.-S. B. Evehe, E.-H. Ba, G. Mombo-Ngoma, A. L. Malisa, R. Ord, W. Inojosa, A. Matondo, D. A. Diallo, W. Mbacham, I. V. van den Broek, T. D. Swarthout, A. Getachew, S. Dejene, M. P. Grobusch, F. Njie, S. Dunyo, M. Kweku, S. Owusu-Agyei, D. Chandramohan, M. Bonnet, J.-P. Guthmann, S. Clarke, K. I. Barnes, E. Streat, S. T. Katokele, P. Uusiku, C. O. Agboghoroma, O. Y. Elegba, B. Cisse, I. E. A-Elbasit, H. A. Giha, S. P. Kachur, C. Lynch, J. B. Rwakimari, P. Chanda, M. Hawela, B. Sharp, I. Naidoo, and C. Roper. 2009. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 6:e1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peek, R., T. Van Gool, D. Panchoe, S. Greve, E. Bus, and L. Resida. 2005. Drug resistance and genetic diversity of Plasmodium falciparum parasites from Suriname. Am. J. Trop. Med. Hyg. 73:833-838. [PubMed] [Google Scholar]
- 25.Plowe, C. V., J. F. Cortese, A. Djimde, O. C. Nwanyanwu, W. M. Watkins, P. A. Winstanley, J. G. Estrada-Franco, R. E. Mollinedo, J. C. Avila, J. L. Cespedes, D. Carter, and O. K. Doumbo. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulphadoxine use and resistance. J. Infect. Dis. 176:1590-1596. [DOI] [PubMed] [Google Scholar]
- 26.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174-1181. [DOI] [PubMed] [Google Scholar]
- 27.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, T. Anderson, C. Roper, R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 28.Walker, A. J., and F. J. Lopez-Antunano. 1968. Response to drugs of South American strains of plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 62:654-667. [DOI] [PubMed] [Google Scholar]
- 29.Wooden, J., S. Kyes, and C. H. Sibley. 1993. PCR and strain identification in Plasmodium falciparum. Parasitol. Today 9:303-305. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, Z., S. M. Griffing, A. M. de Oliveira, A. M. McCollum, W. M. Quezada, N. Arrospide, A. A. Escalante, and V. Udhayakumar. 2008. Decline in sulfadoxine-pyrimethamine-resistant alleles after change in drug policy in the Amazon region of Peru. Antimicrob. Agents Chemother. 52:739-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.