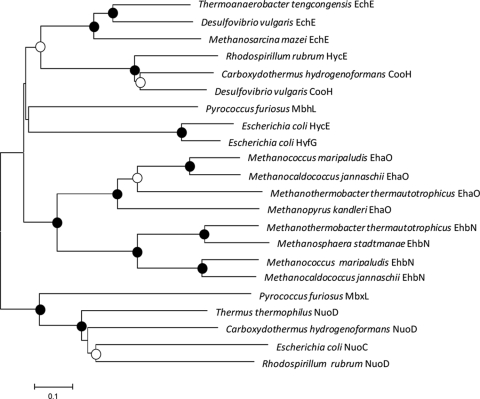
FIG. 2.
Phylogenetic relationships of NiFe large hydrogenase subunits and NADH:ubiquionone oxidoreductase. Amino acid sequence alignments and trees were generated by using the MEGA3.1 program. Alignments were generated by using the CLUSTAL × algorithm. The phylogenetic tree was generated by using the minimum evolutionary distance matrix method, although similar trees were found by using the neighbor-joining and maximum-parsimony methods (data not shown). The bootstrap values are indicated by circles at the branch points: •, >70%; ○, 50 to 70%; unlabeled, <50%. The scale bar is 0.1 expected amino acid substitutions per site. The accession numbers for the amino acid sequences (from the top to the bottom of the tree) are NP_621827, AAS94913, AAM32020, YP_426513, NC_007503, AAS96764, AAL81566, AAC75763, AAC75540, CAF31018, NP_247493, CAB52770, NP_613748, CAB52791, YP_448457, CAF30709, NP_248021, AAL81558, YP_005886, YP_360254, AAC75346, and YP_426645.