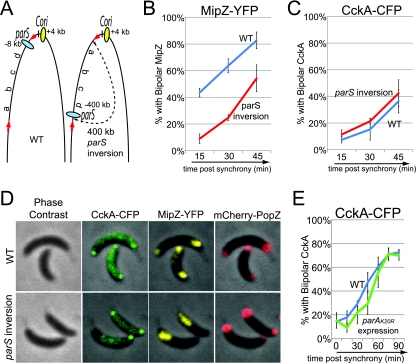
FIG. 4.
DNA segregation is not required for CckA localization. (A) Schematic of the relative positions of chromosomal parS and Cori sequences regions in wild-type cells and in cells with an inversion of a chromosomal region (a 400-kb inverted region is marked between red arrows and contains the parS locus and other regions arbitrarily called a, b, c, and d). (B and C) Time course of the fraction of wild-type (blue line; strain AA877) and parS chromosomal inversion (red line; strain AA878) strains that have bipolar MipZ-YFP, indicating that the origin and parS sequences have been duplicated and segregated to the two cell poles (B) and bipolar CckA-CFP (C) (n = 131 to 704 cells). Swarmer cells were isolated and imaged 15, 30, and 45 min after synchrony. (D) Phase contrast and fluorescence images of CckA-CFP, MipZ-YFP, and mCherry-PopZ localization in wild-type (strain AA877) and parS chromosomal inversion (strain AA878) strains, 45 min after synchronization. (E) Distribution of bipolar CckA-CFP localization in the parA(K20R) expression strain (green line; strain AA843), compared with the wild-type strain (blue line; strain AA841) (n = 121 to 438 cells). Swarmer cells were isolated and imaged every 15 min during 90 min of incubation. For the strain bearing a parA(K20R) allele, cells were grown in the presence of 0.03% xylose for 1 h [to induce parA(K20R)] before the isolation of swarmer cells and then incubated in the presence of 0.03% xylose.