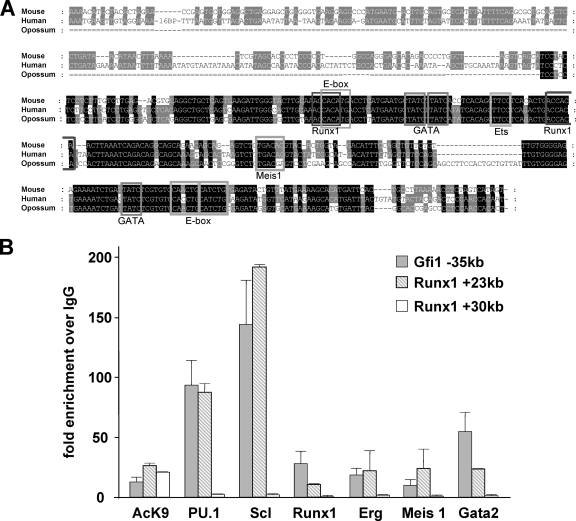
FIG. 6.
Characterization of the Gfi1 kb −35 element identifies several key stem cell transcription factors as upstream regulators. (A) Multispecies alignment of the kb −35 construct highlights the presence of highly conserved transcription factor motifs. (The transcription factor name is identified below the DNA motif.) (B) ChIP assay in HPC-7 cells shows clear binding of Scl, PU.1, Runx1, Erg, Meis1, and Gata2 to the kb −35 construct. All ChIP samples were normalized to IgG and compared to both a positive region (Runx1 +23kb) and a negative region (Runx1 +30kb). The Runx1 kb +23 construct has previously been shown to bind and be activated by the Scl complex of transcription factors, whereas the Runx1 kb +30 construct is known to be acetylated and within a region of open chromatin but is not bound by any transcription factors known to cooperate with Scl (17, 20). Hence, these elements are used as positive and negative regions, respectively.