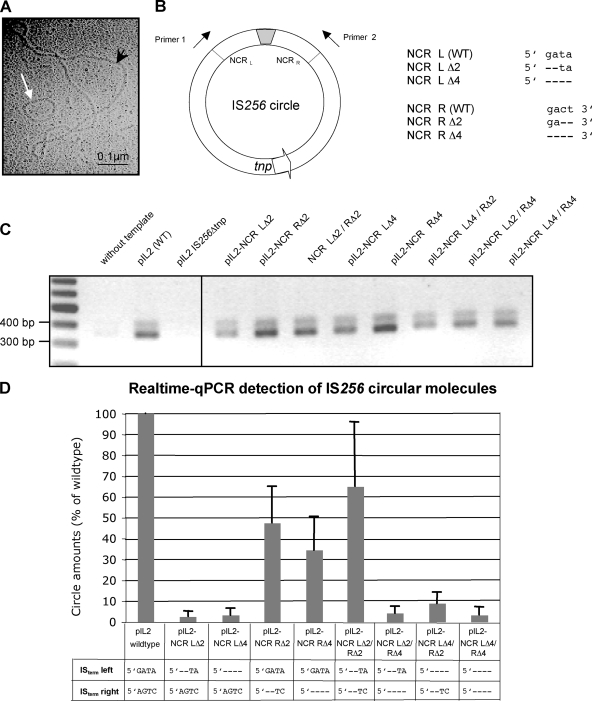
FIG. 5.
Effect of terminal IS deletions on IS256 circle formation. (A) Electron micrograph of an IS256 circular DNA molecule (white arrow). The picture was taken from an E. coli plasmid preparation of vector pIL2 carrying a wild-type IS256 insertion. The black arrowhead marks a 9.3-kb pIL2 vector molecule. (B) Illustration of an IS256 circle with abutted IS ends separated by a short stretch of foreign DNA (gray box). Arrows indicate the direction of primers used for PCR detections of IS256 circles in panels C and D. The orientation of the transposase gene tnp is marked by an arrow. The chart on the right details the nucleotide sequence of the last four nucleotides at the IS256 ends and their deletions introduced into the IS256 copy on vector pIL2, respectively. (C) Agarose gel electrophoresis of IS256 circle-specific PCR products amplified by using primers 1 and 2 and plasmid preparations of pIL2 (as a positive control) and pIL2 IS256Δtnp (as a negative control), as well as pIL2-derived mutants, with deletions in the IS256 termini as templates, respectively. (D) Comparison of the circle amounts detected in pIL2, carrying an IS256 wild-type copy, and various IS termini mutants, respectively, as determined by qPCR.