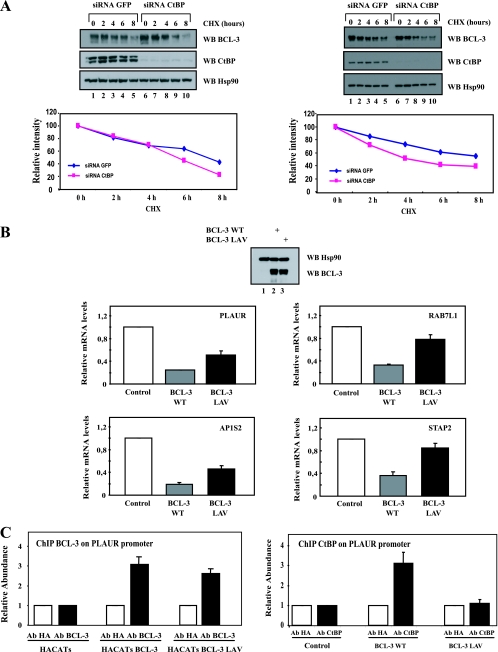
FIG. 5.
CtBP is required for the repressing abilities of BCL-3 and to prevent excessive BCL-3 degradation. (A) CtBP is required for BCL-3 stability. (Top) GFP (negative control)- or CtBP-depleted, BCL-3-expressing 293 (left) or Karpas (right) cells were either left untreated (lanes 1 and 6) or stimulated with cycloheximide (CHX) (50 μg/ml) (lanes 2 to 5 and 7 to 10), and cell extracts were subjected to Western blotting (WB) with antibodies against Hsp90, BCL-3, and CtBP, as indicated. (Bottom) Quantification of BCL-3 levels under control conditions or upon CtBP depletion in BCL-3-expressing 293 cells (left) or in Karpas cells (right). The signal intensity in unstimulated GFP siRNA cells is set to 100%. (B) The N-terminal PVDLR motif is required for the repressing abilities of BCL-3. HaCat cells were infected either with a control lentivirus (negative control) or with a lentivirus expressing WT BCL-3 or BCL-3 LAV. Western blots with antibodies against BCL-3 and Hsp90, performed on extracts from the corresponding experimental conditions in order to ensure comparable levels of WT and mutated BCL-3 products, are shown at the top. Total RNAs from the resulting cells were subjected to real-time PCR in order to assess the mRNA levels of PLAUR, RAB7L1, STAP2, and AP1S2, all genes repressed by WT BCL-3 in transformed keratinocytes (data not shown). The abundance of transcripts in cells infected with the control lentivirus was set to 1, and their levels in cells infected with the other lentivirus were relative to that after normalization with 18S rRNA. Data from three (PLAUR and RAB7L1), four (AP1S2), or five (STAP2) independent experiments (means ± standard deviations) are shown. (C) Chromatin immunoprecipitation assays with an anti-HA (negative control), anti-BCL-3 (left), or anti-CtBP (right) antibody (Ab) were performed using control, WT BCL-3-expressing, or BCL-3 LAV-expressing HaCat cells. Associated DNA was analyzed by real-time PCR using primers derived from the promoter of the PLAUR gene (data not shown). Data from two independent experiments (means ± standard deviations) are shown.