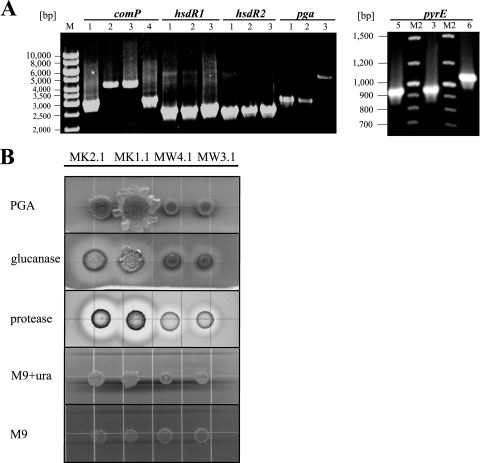
FIG. 2.
Genotypical and phenotypical examination of generated mutants. (A) PCR analysis of chromosomal DNA from B. licheniformis MK2.1 (ΔhsdR1, ΔhsdR2, comP active, ΔpyrE, Δpga) (1), MW4.1 (ΔhsdR1, ΔhsdR2, ΔpyrE, Δpga) (2), MW3.1 (ΔhsdR1, ΔhsdR2, ΔpyrE) (3), F11 (4), MK1.1 (ΔhsdR1, ΔhsdR2, comP active, ΔpyrE) (5), and MW3 (ΔhsdR1, ΔhsdR2) (6). As anticipated, mutants display smaller amplicons than the parental strain. (B) Equal amounts of cells of B. licheniformis MW3.1, MW4.1, MK1.1, and MK2.1 cultures were spotted onto agar plates containing LB only (PGA), LB plus lichenin (glucanase), and M9 plus skim milk (protease), respectively. Additionally, cells were dropped on M9 minimal medium supplemented with uracil (M9+ura) and on M9 minimal medium lacking uracil (M9). Note that the ComP-positive strain displays increased polyglutamate formation in either case.