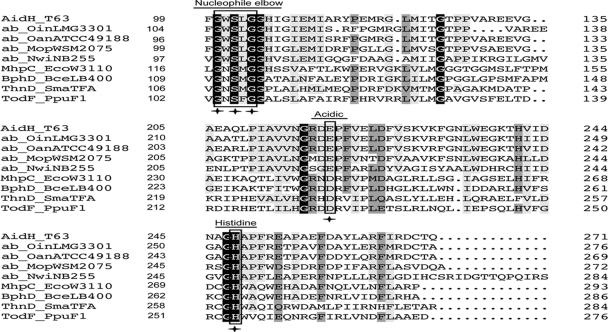
FIG. 2.
Comparison of amino acid sequences of AidH and several alpha/beta-hydrolases. The alignment was generated by DNAMAN. Sections from left to right are the protein, species, and number of amino acids for the gene before the sequences shown. Identities are highlighted in white with a black background, and similarities are shaded gray. The catalytic triad residues are boxed with rectangles. The amino acid residues essential for AHL-degrading activity are indicated by asterisks. AidH_T63, AidH of Ochrobactrum sp. T63 (GenBank accession no. GQ849010); ab_OinLMG3301, alpha/beta-hydrolase fold of O. intermedium (accession no. EEQ96967); ab_OanATCC49188, alpha/beta-hydrolase fold of O. anthropi ATCC 49188 (accession no. YP001369382); ab_MopWSM2075, alpha/beta-hydrolase fold of Mesorhizobium opportunistum WSM2075 (accession no. EEW31886); ab_NwiNB255, alpha/beta-hydrolase fold of Nitrobacter winogradskyi Nb-255 (accession no. YP317111); MhpC_EcoW3110, MhpC from Escherichia coli W3110 (accession no. D86239); BphD_BceLB400, BphD from Burkholderia cepacia LB400 (accession no. X66123); ThnD_SmaTFA, ThnD from Sphingomonas macrogoltabidus TFA (accession no. AF204963); TodF_PpuF1, TodF from Pseudomonas putida F1 (accession no. Y18245).