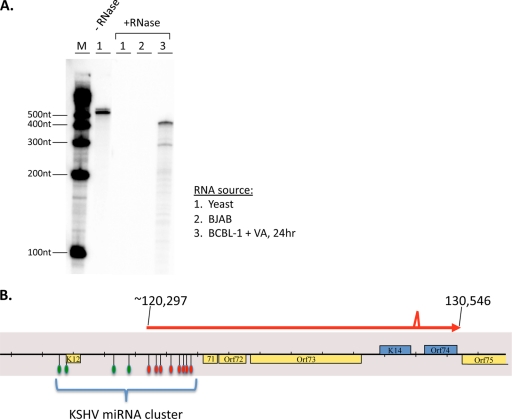
FIG. 6.
Fine-structure mapping of ALT RNA. (A) The RNase protection assay (RPA) was performed on total RNA from yeast cells (lane 1), BJAB cells (lane 2), or VA-induced BCBL-1 cells at 24 h postinduction (+RNase; lane 3) using an undigested riboprobe that spans nucleotide positions 120748 to 120283 (−RNase; lane 1). After hybridization of the radiolabeled riboprobe to the indicated RNAs, the probes were subject to a treatment with RNase A or T1. The protected fragments were analyzed by acrylamide gel electrophoresis. Lane M, molecular size markers. (B) Schematic of ALT RNA transcript structure, based on 5′- and 3′-RACE analyses, RT-PCR analysis, and RPA. The indicated splice site is the same splice site (129219 to 129367) found in the bistronic K14 or vGPCR message. The 3′ end of ALT RNA is coterminal (130546) with the bistronic K14 or vGPCR message. The 5′-RACE clone and the RPA support the conclusion that nt 120297 is the 5′ nucleotide, but other downstream start sites may also exist. Assuming this start site, KSHV pre-miRNAs that are spanned by ALT RNA are indicated by the red symbols, while the pre-miRNAs that are not spanned by the ALT RNA are indicated by the green symbols.