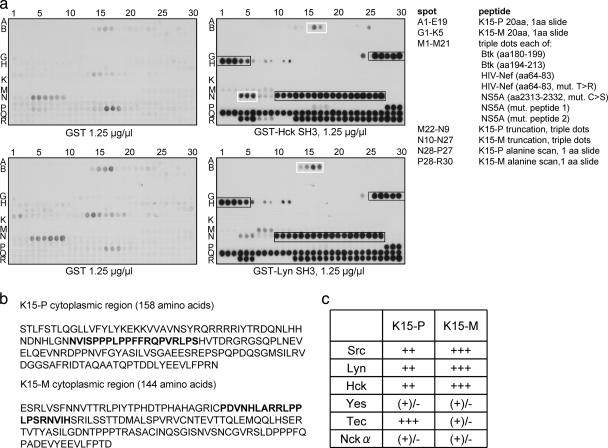
FIG. 1.
The SH3 domains of Hck and Lyn bind to the putative SH3 binding sites of K15-P and K15-M. (a) Overlapping peptides corresponding to the cytoplasmic region of K15-P or K15-M and control peptides were immobilized via their C termini on cellulose membranes and were incubated with recombinant GST alone (left panels) or GST Hck SH3 or GST Lyn SH3 protein (right panels). Spots A1 to E19 (K15-P) and G1 to K5 (K15-M) represent peptides of 20 amino acids, with 1 amino acid slide per peptide to cover the entire cytoplasmic regions of the two K15 proteins. Spots M1 to 21 are peptides derived from proteins known to bind to Hck and Lyn as indicated in the list. Spots M22 to N9 (K15-P) and N10 to 27 (K15-M) represent truncation peptides of the K15 proteins to map the core binding regions within the K15 SH3-B sites (printed in boldface in panel b). Spots N28 to P27 (K15-P) and P28 to R30 represent an alanine scan through the identified K15 motifs (printed in boldface in panel b). (b) Amino acid sequences of the entire cytoplasmic regions of K15-P and K15-M. The identified SH3-B site within the cytoplasmic region of each K15 protein is printed in boldface. (c) Relative binding of individual SH3 domain-displaying phages to GST-K15 (P- and M-type)-coated wells. Wells were coated with recombinantly expressed fusion proteins consisting of GST fused to the cytoplasmic tail of K15-P or K15-M. Individual SH3 domain-displaying phages were incubated with GST fusion protein-coated wells, and the strength of binding was estimated based on the increase (1- to 10,000-fold) of phages bound compared with that in plates coated with plain GST. The table indicates semiquantitative binding strengths of the individual SH3-displaying phages (left vertical column) to GST-K15-P or GST-K15-M fusion proteins (top horizontal column). Binding strengths: +++, strong (>1,000-fold enrichment); ++, good (>100-fold enrichment); +, moderate (>10-fold enrichment); (+)/−, weak/none (<10-fold enrichment).