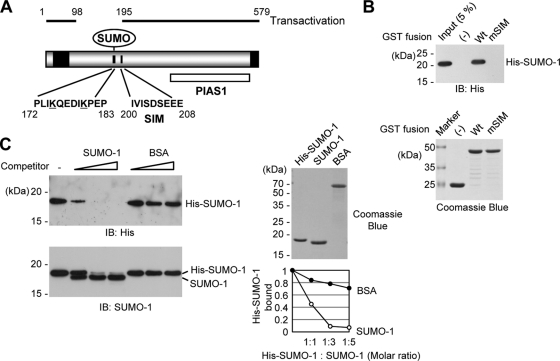
FIG. 1.
Interaction of IE2 with SUMO through a SIM. (A) SUMO-related domain structures of IE2. Two SUMO conjugation sites (K175 and K180), a SIM (between residues 200 and 208), and the protein inhibitor of the activated STAT1 (PIAS1)-binding region (PIAS1) are indicated. Two transactivation domains identified using the GAL4 swap assays (48) are represented as black boxes. The regions of IE2 required for transactivation (residues 1 to 98 and 195 to 579) are also shown. The numbers are amino acid positions of the protein. (B) In vitro binding of IE2 with SUMO-1 through a SIM. The proteins were purified from bacteria and were used in GST pulldown assays. Five micrograms of GST-IE2(135-289) or its SIM mutant version, IE2(135-289/mSIM), was immobilized on glutathione-Sepharose beads and was incubated with 0.5 μg of His-SUMO-1. The bound proteins were fractionated by SDS-PAGE and were detected by immunoblotting with an anti-His Ab (top), and 17% of the GST or GST fusion protein used in the reaction was also fractionated by SDS-PAGE and stained by Coomassie blue (bottom). Five percent of the His-SUMO-1 used in the binding assays is shown as an input control. (C) Competition assays. The in vitro binding assay for which results are shown in panel B was conducted in the presence of increasing amounts of untagged SUMO-1 or BSA (1:1, 1:3, and 1:5 molar ratio with His-SUMO-1). The bead-bound proteins were detected by immunoblotting (IB) with an anti-His Ab (left, top) or with anti-SUMO-1 Ab (left, bottom). Two micrograms of the SUMO-1 and BSA proteins used is shown by Coomassie blue staining (right, top). The relative amounts of His-SUMO-1 bound to IE2 in the presence of competitors were quantified and are shown in the graph (right, bottom).