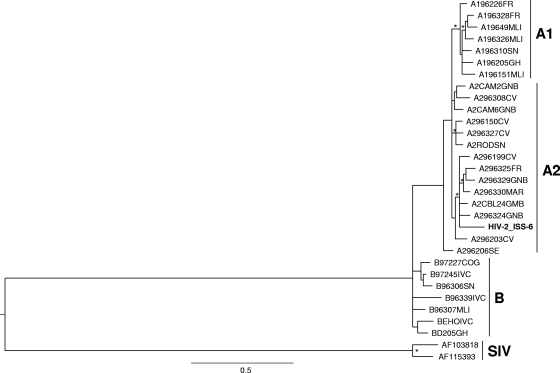
FIG. 1.
Phylogenetic relationships of the HIV-2_ISS-6 isolate (in bold) to representatives of the HIV-2 subtypes A1, A2, B, and SIV (indicated on the right) retrieved from the HIV sequence database (see Materials and Methods). The maximum likelihood (ML) phylogenetic tree was generated from env nucleotide sequences with the Hasegawua, Kishino, and Yano (HKY-85) nucleotide substitution model under gamma distribution (branch lengths were estimated with the best fitting nucleotide substitution model according to a hierarchical likelihood ratio test). The symbol * along a branch represents significant statistical support for the clade subtending that branch (P = 0.001 in the zero-branch-length test; bootstrap support, 75%). The scale bar indicates 0.5% nucleotide sequence divergence. The letters to the right of the sequence name are the country codes for the origins of the patients: FR, France; MLI, Mali; SN, Senegal; GH, Ghana; GNB, Guinea-Bissau; CV, Cape Verde; MAR, Morocco; GMB, Gambia; SE, Sweden; COG, Congo; IVC, Ivory Coast.