Abstract
Gene promoter CpG island hypermethylation is associated with Helicobacter pylori (H. pylori) infection and may be an important initiator of gastric carcinogenesis. To examine factors influencing methylation, we utilized bisulfite Pyrosequencing® technology for quantitative analysis of promoter DNA methylation in RPRM, APC, MGMT and TWIST1 genes using DNA from 86 gastric biopsies from Colombian residents of areas with high and low incidence of gastric cancer. H. pylori colonies were cultured from the same subjects, and gastric pathology was evaluated. Virulence factors cagA (including EPIYA polymorphisms of the 3’ end) and vacA s and m regions were characterized in the H. pylori strains. Using univariate analysis, we found significantly elevated levels of RPRM and TWIST1 promoter DNA methylation in biopsies from residents of the high risk region compared to those from residents of the low risk region. The presence of cagA and vacA s1m1 alleles were independently associated with elevated levels of promoter DNA methylation of RPRM and MGMT. Using multivariate analysis, DNA methylation of RPRM was associated with location of residence, cagA and vacA s1m1 status, and methylation of TWIST1. We conclude that cagA and vacA virulence determinants are significantly associated with quantitative differences in promoter DNA methylation in these populations, but that other as yet undefined factors that differ between the populations may also contribute to variation in methylation status.
Introduction
DNA methylation of promoter CpG islands is an important epigenetic regulatory mechanism for transcription. Methylated cytosines provide binding sites for various methyl-binding proteins, which interact with chromatin remodeling enzymes to stably repress transcription. In normal cells, repetitive elements are inactivated by DNA methylation, while the CpG islands of many housekeeping genes are protected from methylation. Conversely, in cancer cells, repetitive elements lose their methylation, and CpG islands in many promoters become aberrantly methylated. Inactivation of tumor suppressor genes by promoter hypermethylation is considered to be an important contributor to carcinogenesis1.
Gastric cancer is responsible for approximately 800,000 deaths annually worldwide2, and the majority of these occur in the developing world. Infection with Helicobacter pylori (H. pylori) is the major environmental risk factor for gastric cancer3. However, identification of H. pylori infection alone as a risk factor does not simplify the problem of gastric cancer prevention, because approximately half of the world’s population is infected. Most of these persons will have only gastritis, and less than 1% will develop cancer. Global eradication of the infection by antibiotic use is impractical, due to cost and to the problem of generation of antibiotic-resistant strains of H. pylori and other pathogenic bacteria. Homo sapiens and H. pylori have co-evolved over millennia, and infection may produce some benefits for the host, such as reduced rates of asthma and lower risk of esophageal diseases4–6. Therefore identification of more accurate biomarkers for increased risk of gastric cancer could be beneficial in reducing the burden of mortality from this disease.
H. pylori strains vary in their ability to induce and promote gastric cancer. Among the well-established virulence factors is cagA, a marker for a pathogenicity island carried by some strains7, 8. This gene encodes the CagA protein, which is injected into gastric epithelial cells by the type IV Cag secretion system of H. pylori. Once within the cell, CagA becomes tyrosine phosphorylated by Abl and Src family kinases, and then is able to bind to Src Homology 2-containing tyrosine phosphatase (SHP-2) and disrupt cell signaling9. Numerous studies have found an association between the presence of CagA in infecting strains and gastric cancer risk10. Among CagA positive strains, some variants are associated with an even greater risk for gastric cancer. CagA is polymorphic, containing 3 or more EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs within its C-terminus. Among Western H. pylori strains, those with more than 3 EPIYA motifs have greater affinity for SHP-2 and disrupt epithelial cell morphology in vitro11. Notably, persons infected with strains bearing CagA proteins with more than 3 EPIYA motifs are at greater risk for gastric cancer than those harboring strains bearing only 3 motifs12. We previously reported in Colombian populations that persons infected with CagA positive strains with more than 3 EPIYA motifs have more severe gastric precancerous lesions13.
Another important H. pylori virulence factor is vacA, which encodes the vacuolating cytotoxin. This gene is also polymorphic: the sequence encoding the signal peptide is either s1 (with s1a, s1b and s1c subtypes), or s2; the middle portion of the gene is found as m1 or m2 alleles. Strains bearing vacA s1m1 alleles produce more toxin and are associated with more severe pathology than those bearing s2m2 alleles14.
Recent reports indicate that infection with H. pylori is associated with elevated levels of aberrant DNA methylation. Chan et al. found the presence of aberrant methylation of the CDH1 promoter to be associated with the presence of H. pylori in gastric mucosae of dyspeptic patients15. Maekita et al. quantitatively examined methylation levels within promoters of LOX, HAND1, THBD, HRASLS, FLNC, ARC and CDKN2A (p16), and found increased methylation in DNAs from H. pylori-infected subjects compared to uninfected individuals16. Additional affected gene promoters have been identified, including TWIST117–19. Leung et al. found a lower methylation density at the CDH1 promoter after H. pylori eradication20, and an independent study examining H. pylori-associated promoter methylation in 5 genes (CDH1, p16, MLH1, APC, and COX2) similarly reported that aberrant methylation was significantly reduced or completely eliminated one year after H. pylori eradication21. Elevated methylation levels of the FLNC and CDKN2A (p16) genes in gastric mucosae of H. pylori-infected subjects have also been associated with elevated risk for gastric cancer22, 23.
We aimed to determine whether DNA methylation differences could be detected within the context of H. pylori infection in two populations in Colombia. Though situated only 143 miles apart, these two populations differ in gastric cancer incidence by approximately 25-fold (150 vs 6 per 100,000)24. The high risk region is in the rural Andes mountain villages, where the population is supported by agriculture. This population is of mixed Amerindian and Spanish extraction. The low risk region is along the Pacific coast, where the economy is based on fishing. This population is of mixed African and Spanish ancestry. By 5 years of age, 80% of the children in both populations are infected with H. pylori, and infection is acquired at a rate that does not differ significantly between the two populations25. The high risk population has an elevated proportion of subjects infected with more virulent H. pylori strains (CagA positive, vacA s1m1), but this difference is estimated at 24% or less26, 27. These populations provide a useful natural laboratory in which to examine risk factors and potential biomarkers for gastric cancer.
Methods
Human Subjects
The study population consisted of 86 men with dyspeptic symptoms, aged 31 to 60 years old, who underwent upper endoscopy in gastroenterology clinics in two public hospitals between June and September, 2006. Ethics committees of the participating hospitals and of the Universidad del Valle in Cali, Colombia, approved the protocol for this study, and all patients provided informed consent. The subjects were residents of Tuquerres in the Andes Mountains and Tumaco on the Pacific Coast, both in the State of Nariño, Colombia. Exclusion criteria for the study were serious chronic diseases, previous gastrectomy, or ingestion of proton pump inhibitors, H2-receptor antagonists, or antimicrobials in the month prior to the endoscopy. Biopsy samples from the antrum (greater curvature, within 5 cm of the pylorus), incisura angularis (lesser curvature), and corpus (middle anterior wall) were obtained by a single experienced gastroenterologist. One biopsy from each site was fixed in buffered formalin and embedded in paraffin for histopathology. One biopsy from the gastric antrum was frozen in glycerol and thioglycolate for culture of H. pylori organisms. Another biopsy, from the antrum, was frozen without any added solution, for analysis of DNA methylation. Frozen biopsies were shipped on dry ice to Vanderbilt University in Nashville, Tennessee, U.S.A., and stored at −80°C until thawed for culture or methylation analysis.
Histopathology
Four-micron sections were stained with hematoxylin and eosin for diagnosis performed independently by two experienced pathologists (MBP and PC). Discordant diagnoses were reviewed until a consensus was reached. Diagnosis was performed by established guidelines as normal, non-atrophic gastritis (NAG), multifocal atrophic gastritis without intestinal metaplasia (MAG), intestinal metaplasia (IM), or dysplasia (DYS) 28, 29. Pathologists evaluated diagnoses blinded to residence of subjects.
H. pylori culture and genotyping
One antral and one corpus biopsy per person were homogenized under sterile conditions and cultured on selective Trypticase soy agar with 5% sheep blood and vancomycin (20 µg/ml), bacitracin (200 µg/ml), nalidixic acid (10 µg/ml) and amphotericin B (2 µg/ml), (all from Sigma, St Louis, MO), as previously described13. Small gray translucent colonies characteristic of H. pylori appeared after 4 to 8 days. Identity of H. pylori was confirmed by morphology, Gram stains and assays for oxidase and urease. Pellets from three single colonies per biopsy were frozen for analysis. DNA was isolated with either a Puregene kit (Qiagen) or by proteinase K digestion overnight followed by phenol / chloroform extraction. In preliminary experiments, 2 to 5 colonies per person were analyzed from 28 subjects by DNA fingerprinting analysis to evaluate the frequency of mixed infections in the colonies harvested from the antral and corpus biopsies30. In all 28 subjects, all colonies from the same person produced identical DNA amplification patterns, indicating that there were no mixed infections in the strains harvested from a single individual. Subsequently, all analyses for H. pylori virulence markers were performed on DNA from a single colony per subject, isolated from the antral biopsy.
DNA from one colony per participant was genotyped for cagA and vacA s and m regions. Analysis of cagA, including the polymorphic 3’ end, was previously described from H. pylori strains from this study population13. The vacA s region of each strain was amplified by primers VA1F14 and Vac785 (R: AATACGCTCCCACGTATTGC), using an annealing temperature of 52°C. PCR products were treated with ExoSAP-IT (USB, Cleveland, OH) and sequenced using BigDye 3.1 terminator chemistry with an ABI 3130xl Genetic Analyzer. Two strains were genotyped for the vacA s region by PCR alone, using primers VA1F and VA1R14 followed by electrophoresis in a 1.5% agarose gel, discriminating a 259 bp fragment for s1 or a 286 bp fragment for s2. The vacA m region was defined by multiplex PCR using primers M1F2, M1R2, M2F1, and M2R131. Primer sequences and details of amplifications are described in Sicinschi et al27.
Methylation Analysis
We used a Pyrosequencing® technique, which allows independent quantitative determination of methylation at selected CpG sites in the gene promoter. To obtain material for the analyses, gastric antral biopsies, one intact biopsy per person, were homogenized using disposable plastic pestles and digested overnight with proteinase K. DNA was isolated by phenol / chloroform extraction and quantitated with a Nanodrop instrument (Thermo Scientific, Wilmington, DE); 0.5 to 2 ug of DNA per biopsy was bisulfite modified, using a Zymo EZ Methylation Direct kit (Zymo Research Corp., Orange, CA). Modified DNA (20 ng per reaction) was amplified by PCR, using 0.2 uM of each primer, 2 Units of hot start Taq DNA polymerase, and 0.2 mM of each dNTP per reaction. Primer sequences and their location are listed in Table 1. Cycling programs were 95°C for 15 min., then 48 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, followed by a 5 min. incubation at 72°C. PCR products were examined following gel electrophoresis in 1.5% agarose, in order to confirm that a single band was obtained. In preparation for Pyrosequencing, the biotinylated strand of each PCR product was isolated from 10 to 20 ul of each PCR product using a Vacuum Prep Tool (Qiagen Inc., Valencia, CA.), according to the manufacturer’s protocol. Pyrosequencing reactions were performed in a PyroMark MD Pyrosequencing instrument (Qiagen), in the presence of 500 nM sequencing primer, following the manufacturer’s recommendations. The positive control for methylation was methylated HeLa DNA (New England Biolabs, Ipswich, MA); the negative control was normal human blood DNA (Promega Corp., Madison, WI). Positive and negative controls were run with each experiment. Pyrosequencing assays contained a control for incomplete bisulfite modification. Methylation values used for analysis were the mean of the percent methylation measured for the first 3 potential methylation sites following the sequencing primer.
Table 1.
Primer sequences for Pyrosequencing
|
APC Promoter 1A |
F: GGGGTTAGGGTTAGGTAGGTT R: biotin-ACTACACCAATACAACCACATATC Seq: GAGAGAAGTAGTTGTGTAAT |
*chr 5: 112,101,273-112,101,293 antisense 112,101,444-112,101,472 112,101,348-112,101,367 |
** Industry source |
| MGMT | F: TGGTTTGGGGGTTTTTGA | chr 10, 131,154,890-131,154,907 | This study |
| R: biotin-CCTTTTCCTATCACAAAAATAATC | antisense 131,155,065-131,155,088 | ||
| Seq: ATTAGGAGGGGAGAGAT | 131,154,921-131,154,937 | ||
| RPRM | F: GGTTATTAAGGAAGTTTGGGTGTA | chr2:154,043,215-154,043,238 | This study |
| R: biotin-AACCCCACACCTATTCCTCC | antisense 154,043,387-154,043,406 | ||
| Seq: TTGTTTAGGGTAGGATTTAT | 154,043,306-154,043,325 | ||
| TWIST1 | F: GTTAAGTGAGGTGGGAAGGTTGA | chr7:19,122,688-19,122,710 | This study |
| R: biotin-CCCACCCCCTCAACAAAAC | antisense 19,122,830-19,122,848 | ||
| Seq: GGAGAGGGGAGGAAA | 19,122,721- 19,122,735 |
Position indicates equivalent location of primer (in unmodified sequence) in the March 2006 human reference sequence (NCBI Build 36).
assay designed by QIAGEN AB, formerly Biotage AB, as published in PyroMark™ Assay Database at www.pyrosequencing.com
Statistical Analysis
Age, histological diagnoses, levels of methylation at the 4 gene promoters, H. pylori virulence genes, and number of EPIYA motifs were compared in a univariate fashion by risk area using χ2, Fisher's exact, Wilcoxon/Mann Whitney, Spearman, and Student’s t tests, as appropriate. Mathematical transformations of the 4 levels of methylation were evaluated to reduce data skewness. H. pylori status was analyzed several ways combining cagA and vacA genotyping. In order to assess the degree to which various factors explained levels of methylation, multivariate generalized linear models were used. For continuous and binary variables, the coefficients represent the average difference in percentage of methylation for a one unit change in the predictor (i.e. for every year of age, or low versus high risk area). In the case of H. pylori genotypes, the coefficient represents the average difference in percentage of methylation associated with being a part of the specified category (cagA and vacA s1m1 or other genotypes) as compared with the reference group (uninfected). Diagnosis was considered as an ordinal variable (0=Normal/NAG, 1=MAG, and 2=IM/DYS), for purposes of estimating the p-values in the test of linear trend (p-trend).
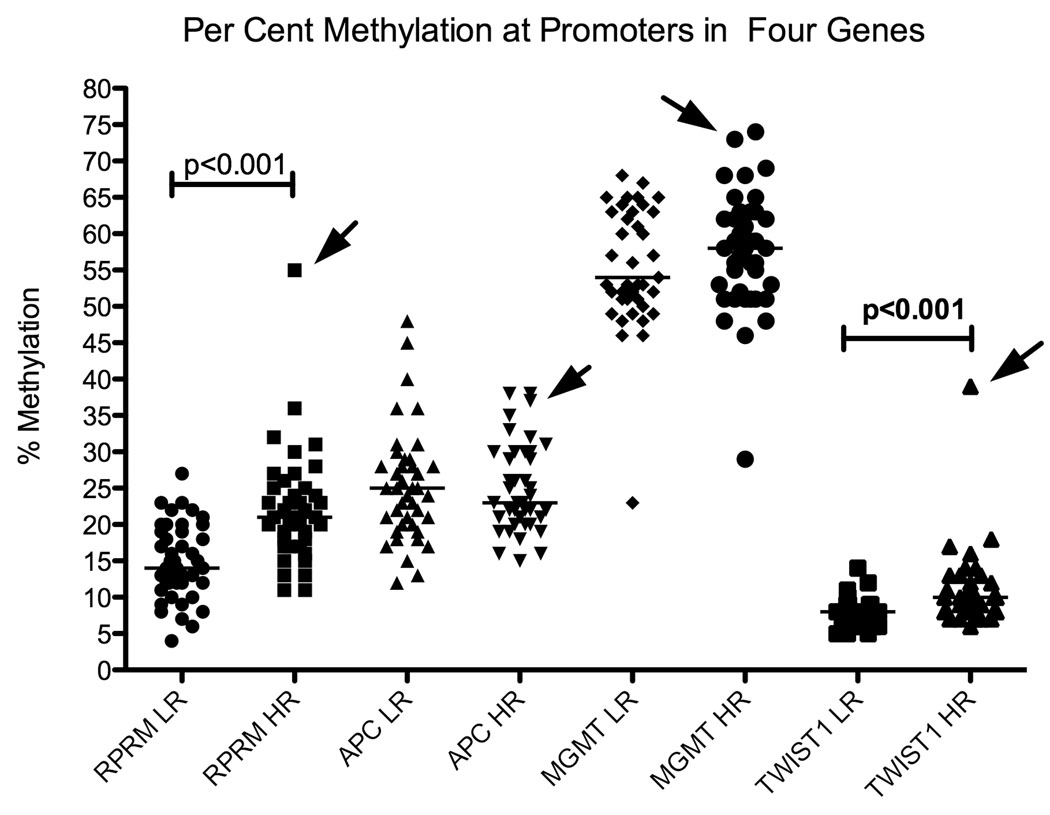
Regression analyses were individually applied to original and mathematically transformed variables. Since the results were similar, only those results based on the original variables are presented. Data were analyzed using the software package Stata 10.0 (Stata Corporation, College Station, TX). Prism 5.0b (GraphPad Software, La Jolla, CA) was used to generate Figure 1.
Figure 1.
Scatter plots indicate the percent methylation of each gene, measured from DNA from residents of low risk (LR) and high risk (HR) areas. Arrows indicate the points representing the outlier subject, whose gastric histology showed widespread intestinal metaplasia and focal areas of indefinite dysplasia.
A kappa coefficient of reliability was run on 64 replicates to assess reproducibility of the Pyrosequencing results. This analysis produced a kappa statistic of 0.29 (95% CI=0.27 – 0.37, bias corrected), with complete agreement of 31% of samples, a standard error of 0.023, and a correlation coefficient of 0.983 (p<0.0001).
Results
Subjects and their Infecting H. pylori Strains
Table 2 shows the description of the subjects from the high and low risk populations and their infecting H. pylori strains. Age distributions and prevalence of H. pylori infection were similar in the two groups, but the high risk population had more advanced gastric precancerous lesions (p=0.021). Among the 64 strains that were analyzed for vacA s region by sequencing, all were s1b. Among the cagA positive strains, most had EPIYA patterns of ABC (66% of the cagA positive strains from the low risk region, and 57% of those from the high risk region). This difference was not significant. Of the remaining cagA positive strains, most were ABCC, but some variants were found: an AC strain in the low risk region, and one each of ABCCC, ABBCC, ABBC, ABCBC and BCC in the high risk region32.
Table 2.
Study population and Characteristics of Infecting H. pylori Strains
| Variables | Low risk for Gastric Cancer n=45 |
High risk for Gastric Cancer n=41 |
Two-tailed p-value * |
|---|---|---|---|
| Age in years, mean (SD) | 47.5(6.0) | 48.9(5.3) | 0.257 |
| cagA assessment by PCR, n(%) | |||
| Uninfected | 7(15.6) | 2(4.9) | 0.209 |
| Negative | 6(13.3) | 4(9.7) | |
| Positive | 32(71.1) | 35(85.4) | |
| H. pylori genotypes, n(%) | |||
| Uninfected | 7(15.6) | 2(4.9) | 0.079 |
| cagA positive, vacA s1m1** | 29(64.4) | 35(85.4) | |
| Other genotypes | 9 (20.0) | 4 (9.7) | |
| H. pylori genotypes, n(%) | |||
| Uninfected | 7(15.6) | 2(4.9) | 0.128 |
| cagA negative | 6(13.3) | 4(9.7) | |
| cagA positive, 3 EPIYAs | 25(55.6) | 21(51.2) | |
| cagA positive, >3 EPIYAs | 7(15.5) | 14(34.2) | |
| H. pylori genotypes, n(%) | |||
| uninfected | 7(15.6) | 2(4.9) | 0.076 |
| cagA positive, (3 EPIYAs), vacA s1m1* | 7(15.6) | 14(34.2) | |
| cagA positive (>3 EPIYAs) vacA s1m1* | 22(48.8) | 21(51.2) | |
| other genotypes | 9(20.0) | 4(9.7) | |
| Histopathological diagnosis§ , n(%) | |||
| Normal/NAG | 27(61.4) | 13(31.7) | 0.021 |
| MAG | 6(13.6) | 8(19.5) | |
| IM/Dysplasia | 11(25.0) | 20(48.8) | |
| % methylation, median (interquartile range) | |||
| RPRM | 14(7) | 21(7.5) | <0.001 |
| APC | 25(8) | 23(9) | 0.9158 |
| MGMT | 54(12) | 58(11) | 0.3788 |
| TWIST1 | 8 (2) | 10(4) | <0.001 |
| Mathematically transformed values of % methylation, mean (SD) |
|||
| RPRM (logarithm) | 2.6(0.4) | 3.0(0.3) | <0.001 |
| APC (logarithm) | 3.2(0.3) | 3.2(0.2) | 0.9460 |
| MGMT(square-root) | 3160(848.4) | 3369(906.6) | 0.2869 |
| TWIST1(inverse) | 0.10 (0.03) | 0.13 (0.03) | <0.001 |
Abbreviations: SD= standard deviation; NAG: non-atrophic gastritis; MAG: multifocal atrophic gastritis; IM: intestinal metaplasia.
p-value from χ2, Fisher's exact, t test or Wilcoxon/Mann-Whitney test as appropriate.
This category includes a subject with a strain of H. pylori that was cagA positive and vacA s1, but which repeatedly failed PCR for vacA m.
A subject with gastritis in which we could not assess atrophy was excluded from this analysis.
Unadjusted Analysis
In univariate analysis, levels of methylation (either medians or means) in the RPRM and TWIST1 promoters were significantly associated with the area of residence of the subject, with more methylation being found in the DNA from subjects in the high risk area. No significant difference was detected for the APC or MGMT promoters.
Considering results from the entire set of DNA samples, generalized unadjusted linear regression models showed that levels of methylation in the RPRM and MGMT promoters were positively and significantly associated with histological diagnoses (Coefficient: 2.1; p-trend =0.018 for RPRM; Coefficient: 3.0; p-trend=0.003 for MGMT). Coefficients indicate that as lesions progressed, higher levels of methylation were observed. In the small number of uninfected patients, median per cent methylation was 8% for RPRM (n=4), 38% for APC (n=3), 39% for MGMT (n=4) and 6.5% for TWIST1 (n=4). Regarding H. pylori infection, the presence of cagA and vacA s1m1 alleles was associated with elevated levels of methylation of RPRM and MGMT. Interestingly, the association with APC was inverse, such that more virulent H. pylori strains were associated with less methylation of this gene. Age was not associated with levels of methylation.
Multivariate Analysis
Multivariate regression models incorporated effects of geographic area, diagnosis, age, and genotypes of the infecting H. pylori strain (classified as uninfected, cagA positive, vacA s1m1 and all other genotypes; Table 3). Because alternative grouping of variables of H. pylori status, including numbers of EPIYA motifs, or analysis by the presence of cagA alone, did not improve the explanatory capacity of the models, and led to similar results, parsimonious models with the highest adjusted R2 (coefficient of determination) are presented. In these models, the observed mean differences between area and RPRM or TWIST1 remained statistically significant, as did the differences between H. pylori virulence genes and RPRM, MGMT, or APC. Adjustment for covariates significantly attenuated the differences between histological diagnoses and levels of methylation of RPRM and MGMT. Age did not have any significant effect on the levels of methylation.
Table 3.
Multivariate Linear Regression Analysis.
| Variables | Gene |
|||||||
|---|---|---|---|---|---|---|---|---|
| RPRM (n=84) | APC (n=84) | MGMT (n=80) | TWIST1 (n=82) | |||||
| Coefficient (SE) | p-value | Coefficient (SE) | p-value | Coefficient (SE) | p-value | Coefficient (SE) | p-value | |
| Area | ||||||||
| Low risk for Gastric Cancer | 0 | 0 | 0 | 0 | ||||
| High risk for Gastric Cancer | 6.4 (1.4) | <0.001 | 1.0 (1.5) | 0.511 | −1.5 (1.8) | 0.383 | 3.1 (0.9) | 0.001 |
| Diagnosis | ||||||||
| Normal/NAG | 0 | 0 | 0 | 0 | ||||
| MAG | 1.1 (2.0) | 0.583 | −0.6 (2.1) | 0.766 | 1.3 (2.5) | 0.594 | −0.4 (1.4) | 0.746 |
| IM/DYS | 0.2 (1.7) | 0.926 | −1.4 (1.7) | 0.431 | 3.3 (2.1) | 0.124 | 0.4 (1.1) | 0.718 |
| H. pylori genotypes | ||||||||
| uninfected | 0 | 0 | 0 | 0 | ||||
| other genotypes | 5.6 (2.6) | 0.040 | −9.3 (2.9) | 0.002 | 5.3 (3.1) | 0.104 | 0.6 (1.7) | 0.746 |
| cagA positive, vacA s1m1* | 8.3 (2.4) | 0.001 | −9.9 (2.6) | <0.001 | 11.2 (2.9) | <0.001 | 1.8 (1.6) | 0.244 |
| Age in years | −0.1 (0.1) | 0.271 | 0.1 (0.1) | 0.403 | −0.1 (0.1) | 0.598 | −0.1 (0.1) | 0.462 |
| Adjusted R-squared | 0.3356 | 0.1267 | 0.2216 | 0.1281 | ||||
Abbreviations: SE= standard error; NAG: non-atrophic gastritis; MAG: multifocal atrophic gastritis; IM: intestinal metaplasia.
This category includes a subject with a strain of H. pylori that was cagA positive and vacA s1, but which repeatedly failed PCR for vacA m.
Because significant correlations exist between TWIST1 (correlation coefficient rho=0.58; p<0.001) and MGMT (rho=0.39; p=0.001) with RPRM, the combined effect of methylation in these genes was simultaneously evaluated in a regression model using methylation of RPRM as outcome and including the covariates from the multivariate model. When TWIST1 and MGMT were included as covariates, TWIST1 was significantly associated with RPRM (coefficient, 1.04; p<0.0001) and MGMT showed a trend toward significance (Coefficient, 0.14; p=0.078). All effects were independent, as there was no significant interaction or effect modification between any of the covariates in the multivariate model.
An Interesting Outlier
One biopsy from a resident of the high risk region was a notable outlier. DNA from this biopsy showed elevated methylation at all four regions examined, for RPRM (55% methylation, compared to a median of 21%, 95% CI: 19–23%), for TWIST1 (38% methylation, compared to a median of 10%, 95% CI: 8–11%), for APC (37%, compared to a median of 23%, 95% CI: 22–27%) and MGMT (73%, compared to a median of 58%, 95% CI: 55–61%; Figure 1). Histopathological analysis of paraffin-embedded biopsies of the same subject showed extensive intestinal metaplasia in all three sites examined (antrum, corpus and incisura angularis) and focal areas of indefinite dysplasia in antrum and corpus.
Discussion
In an attempt to understand mechanisms that regulate progression towards gastric cancer, we compared two populations that differ greatly in the incidence of this disease, although they have equally high prevalence of H. pylori infection26, 27. As expected, the two populations differed in diagnosis, with more severe lesions occurring more frequently in the high risk region. We detected a trend for the presence of the more virulent cagA positive, vacA s1m1 strains more often in the high risk region, consistent with our findings in prior studies with different subjects from these populations26, 27. The quantitative method used to measure DNA methylation allowed us to detect differences in levels of methylation previously missed by earlier methods, such as methylation-specific PCR, which reveals only that methylation is present, without quantitating it. In our study, low levels of methylation present in uninfected patients could be measured; this methylation may occur due to environmental stressors not examined in our analysis. Univariate analysis comparing methylation levels of tissues representing the two populations showed differences in methylation of two genes: RPRM and TWIST1. When we examined several possible covariates of methylation, using multivariate analysis, geographic area remained as an independent effect, with more methylation in RPRM and TWIST1 in the high risk region.
Because many epidemiological studies have shown an association of cagA and vacA s1m1 with gastric cancer risk, we sought to determine if these virulence factors were associated with promoter methylation in the four genes examined. For RPRM and MGMT, an independent effect of cagA and vacA s1m1 on methylation levels was detected. In contrast, for APC, decreased levels of promoter methylation were associated with more virulent H. pylori genotypes. Unadjusted analyses showed that levels of methylation in the RPRM and MGMT promoters were significantly associated with the histological diagnoses, but adjustment for genotypes of the infecting H. pylori strains significantly attenuated the associations. This attenuation is likely due to the association between H. pylori infection and diagnosis of more severe lesions. Our study also demonstrated differential methylation of RPRM and TWIST1 promoters by geographic area, even after adjustment for histological diagnosis and virulence determinants. These results indicate that other virulence determinants of the infecting strains are likely to be responsible for those differences, and/or that additional features distinguishing the two populations, such as diet or genetic features of the host, are important in differentiating risk.
In a prior examination of promoter methylation status and H. pylori virulence determinants, Watanabe et al. examined methylation of MINT25, RORA, GDNF, ADAM23, PRDM5, and MLF1 in gastric washings from Korean subjects with and without gastric cancer33. These investigators tested for the presence of cagA in infecting strains and found no association with methylation. A difference from our study, besides the ethnicity of the subjects, is that Watanabe et al. assayed cagA by PCR on DNA from gastric washings, in contrast to our method of PCR using DNA from cultured bacteria. Culture is likely to be a more sensitive method for detection of H. pylori and its virulence determinants. In addition, as gastric lesions become more advanced, H. pylori infection is often lost, so that evaluation of H. pylori and its virulence determinants in gastric cancers may underrepresent the infection originally present34. Thus our study on premalignant lesions may have been more likely to detect infections originally present.
Park et al. examined methylation levels in a set of genes by quantitative methods, comparing premalignant gastric tissues and gastric cancers, with and without H. pylori infection35, and found significant differences between infected and uninfected subjects, in the DNA samples from gastritis, but not from intestinal metaplasia, adenomas, or gastric cancers. While some studies have found H. pylori-related methylation to be associated with age19, 36, others15, 37, including the current study, have found no significant associations. However, the age range of our subjects was 29 years, and it is possible that with a broader age range, a significant association might be detected.
The genes we chose to study have been previously noted to be associated with development of gastric cancer. One such gene was RPRM, a TP53-dependent mediator of the G2/M cell cycle checkpoint38. RPRM is a tumor suppressor, which is inactivated by promoter methylation in a variety of human tumors, including gastric adenocarcinoma39. Notably, RPRM promoter methylation has been reported in premalignant conditions such as Barrett’s esophagus40 and non-malignant gastric epithelia39. Aberrant methylation of RPRM may be detected by methylation-specific PCR (MSP) in plasma of gastric cancer patients, in contrast to plasma of control subjects, suggesting the potential of RPRM as a biomarker for early gastric cancer41. Our results also support the identification of this gene as one that may be altered early in the progression towards gastric cancer.
Another gene we examined was the APC (adenomatous polyposis coli) gene, which encodes a component of a multiprotein complex that inactivates β-catenin in the Wnt signaling pathway. A critical tumor suppressor in the gastrointestinal tract, APC is expressed in the stomach as two isoforms originating from promoters 1A and 1B42. Methylation in gastric tissue occurs predominantly in promoter 1A, and for this reason, this promoter is frequently examined in studies of hypermethylation in gastric cancer43, 44. However, in contrast to the colon, methylation at promoter 1A in gastric mucosae is not predominantly tumor related, but is frequently found in non-malignant gastric mucosae45, as we also observed. Promoter 1B is not methylated, and gene expression studies have shown that transcription of APC in the stomach is regulated primarily from this promoter45, 46. Consequently methylation at promoter 1A in gastric mucosae is likely to be a passenger, rather than a driver of carcinogenesis. Among the four genes we examined, after adjustment for diagnosis, it is notable that only the APC promoter showed a decrease in methylation in DNAs from subjects from the high risk region.
MGMT encodes O-6-methylguanine-DNA methyltransferase (also called O6-alkylguanine-DNA alkyltransferase), a DNA repair enzyme that removes alkylation adducts from guanine. MGMT must be constantly renewed, as part of defense against mutagens. MGMT promoter hypermethylation is associated with a wide variety of tumors, including gastric cancers47. Our assays detected a relatively high level of MGMT promoter methylation in most of the gastric biopsies. Additional studies will be necessary to determine how these elevated levels are related to MGMT gene expression.
TWIST1 encodes a basic-helix-loop-helix transcription factor called Twist1 that regulates metastasis48 and epithelial-to-mesenchymal transition49. TWIST1 represses transcription of E-cadherin and TIMP1 (an inhibitor of matrix metalloproteinases)50 and promotes migration in neoplastic cells51, 52. Although the Twist1 protein may have tumor- or metastasis-promoting effects, paradoxically, hypermethylated TWIST1 is reported in cancers of the stomach and other tumor types49, 53. All of our Colombian subjects, with the exception of one outlier, had relatively low levels of TWIST1 hypermethylation.
Limitations of our study include the fact that we examined DNA from biopsies containing premalignant lesions, instead of DNA from gastric cancers. Our goal was to investigate early events in gastric carcinogenesis, and these events may not all lead to cancer. Another limitation is the use of entire biopsies for our DNA preparation, so that mixed cell types are included. Although most of the cells contributing DNA were gastric epithelial cells, other cells including stromal and inflammatory cells may have contributed to the differences in methylation levels that we measured. Additional studies employing microdissection will be useful to confirm identity of the cell type demonstrating alterations in methylation levels.
A topic of intense investigation is the mechanism(s) by which promoter methylation becomes dysregulated during the development of cancer, and how inflammation may promote this process. In our study, we found that two of the four genes we examined (RPRM and TWIST1) increased together in levels of methylation, and a third (MGMT) trended toward such an increase, consistent with a common or related mechanism affecting most of the genes we studied. Results from an outlier subject, whose biopsy showed high or remarkably elevated promoter methylation in all four genes examined, are consistent with the existence of a global methylation defect that can occur early in the stages of carcinogenesis. Although this widespread hypermethylation was observed in only one subject, we expect progression of H. pylori-related disease to occur in only a small minority of infected persons, and therefore events occurring within the gastric mucosae of these rare individuals provoke great interest. Analysis of DNA methylation of additional genes and the effect of that methylation on gene transcription will be necessary to clarify early events in H. pylori-related gastric cancer development.
The mechanisms by which virulent H. pylori infection may influence promoter hypermethylation remain obscure, but several hypotheses which involve enhancement of DNA methyltransferase activity have been proposed. H. pylori infection promotes gastric epithelial cell proliferation and increased production of epidermal growth factor (EGF) and its receptor54; the EGF receptor has greater activity with the more virulent cagA positive strains55. EGF administered to gastric epithelial cells in culture increases DNA methyltransferase activity56.
Another possible mechanism by which H. pylori can promote hypermethylation involves the effect of nitric oxide on DNA methyltransferase. Katayama et al. noted that the RUNX3 gene promoter in the human gastric cancer cell line MKN45 can become methylated by co-culture with H. pylori and macrophages, and this effect can be replicated by substituting nitric oxide (NO) for the bacteria and macrophages57. Inhibition of NO synthesis in the co-culture reversed the methylation. Hmadcha et al reported that treatment of an insulinoma cell line by IL-1β or NO could inactivate the FMR1 and HPRT genes by hypermethylation58. This effect was apparently mediated by enhancement of DNA methyltransferase activity, without an increase in transcription of the methyltransferase gene.
In conclusion, RPRM and TWIST1 promoter DNA methylation were found to be elevated in gastric biopsies from residents of the high risk region compared to those from residents of the low risk region in Colombia. Virulence genes cagA and vacA s1m1 in the infecting H. pylori strains were independently associated with elevated levels of methylation of RPRM and MGMT. Alterations in DNA methylation are early changes in the progression of premalignant lesions and have the potential to serve as biomarkers for gastric cancer risk.
Acknowledgements
Acknowledgement of Financial Support: This study was supported by 1 UL1 RR024975 from NCRR/NIH (CTSA), grants P01 CA028842, P50 CA095103, R01 CA77955, R01 CA93999 and P01 CA116087 from the National Cancer Institute, and P30DK058404, R01 DK053620, and DK058587. The content presented here is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
We wish to acknowledge Ms. Jacki Huckins (Qiagen) for assistance in assay design for the RPRM Pyrosequencing assay.
Bibliography
- 1.Vogiatzi P, Vindigni C, Roviello F, Renieri A, Giordano A. Deciphering the underlying genetic and epigenetic events leading to gastric carcinogenesis. J Cell Physiol. 2007;211:287–295. doi: 10.1002/jcp.20982. [DOI] [PubMed] [Google Scholar]
- 2.Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, Thun MJ. Global Cancer Facts & Figures 2007. American Cancer Society. 2007 [Google Scholar]
- 3.Webb PM, Law M, Varghese C, Forman D, Yuan JM, Yu M, Ross R, Limberg PJ, Mark SD, Taylor PR, Dawsey SM, Qiao YL, et al. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut. 2001;49:347–353. doi: 10.1136/gut.49.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167:821–827. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 5.Vaezi MF, Falk GW, Peek RM, Vicari JJ, Goldblum JR, Perez-Perez GI, Rice TW, Blaser MJ, Richter JE. CagA-positive strains of Helicobacter pylori may protect against Barrett's esophagus. Am.J.Gastroenterol. 2000;95:2206–2211. doi: 10.1111/j.1572-0241.2000.02305.x. [DOI] [PubMed] [Google Scholar]
- 6.Loffeld RJLF, Werdmuller BFM, Kuster JG, Pérez-Pérez GI, Blaser MJ, Kuipers EJ. Colonization with cagA-positive Helicobacter pylori strains inversely associated with reflux esophagitis and Barrett's esophagus. Digestion. 2000;62:95–99. doi: 10.1159/000007801. [DOI] [PubMed] [Google Scholar]
- 7.Tummuru MKR, Cover TL, Blaser MJ. Cloning and expression of a high-molecular mass major antigen of Helicobacter pylori: Evidence of linkage to cytotoxin production. Inf Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Ziang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Nat Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr Opin Microbiol. 2008;11:30–37. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125:1636–1644. doi: 10.1053/j.gastro.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S, Azuma T, Hatakeyama M. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology. 2006;130:1181–1190. doi: 10.1053/j.gastro.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 12.Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sicinschi LA, Correa P, Peek RM, Jr, Camargo CM, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Mera R, Delgado AG, Bravo LE, Schneider BG. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2009 doi: 10.1111/j.1469-0691.2009.02811.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atherton JC, Cao P, Peek RM. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 15.Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502–506. doi: 10.1136/gut.52.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 17.Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161–170. doi: 10.1038/labinvest.3700707. [DOI] [PubMed] [Google Scholar]
- 18.Nakajima T, Yamashita S, Maekita T, Niwa T, Nakazawa K, Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer. 2009;124:905–910. doi: 10.1002/ijc.24018. [DOI] [PubMed] [Google Scholar]
- 19.Kitajima Y, Ohtaka K, Mitsuno M, Tanaka M, Sato S, Nakafusa Y, Miyazaki K. Helicobacter pylori infection is an independent risk factor for Runx3 methylation in gastric cancer. Oncol Rep. 2008;19:197–202. [PubMed] [Google Scholar]
- 20.Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, Cheng VY, Ng EK, Sung JJ. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216–3221. doi: 10.1158/1078-0432.CCR-05-2442. [DOI] [PubMed] [Google Scholar]
- 21.Perri F, Cotugno R, Piepoli A, Merla A, Quitadamo M, Gentile A, Pilotto A, Annese V, Andriulli A. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol. 2007;102:1361–1371. doi: 10.1111/j.1572-0241.2007.01284.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima T, Maekita T, Oda I, Gotoda T, Yamamoto S, Umemura S, Ichinose M, Sugimura T, Ushijima T, Saito D. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317–2321. doi: 10.1158/1055-9965.EPI-06-0436. [DOI] [PubMed] [Google Scholar]
- 23.Tahara T, Arisawa T, Shibata T, Wang FY, Nakamura M, Sakata M, Nagasaka M, Takagi T, Kamiya Y, Fujita H, Nakamura M, Hasegawa S, et al. Risk prediction of gastric cancer by analysis of aberrant DNA methylation in non-neoplastic gastric epithelium. Digestion. 2007;75:54–61. doi: 10.1159/000101775. [DOI] [PubMed] [Google Scholar]
- 24.Correa P, Cuello C, Duque E, Burbano LC, Garcia FT, Bolanos O, Brown C, Haenszel W. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst. 1976;57:1027–1035. doi: 10.1093/jnci/57.5.1027. [DOI] [PubMed] [Google Scholar]
- 25.Camargo MC, Yepez MC, Ceron C, Guerrero N, Bravo LE, Correa P, Fontham ETH. Age at acquisition of Helicobacter pylori infection: Comparison of two areas with contrasting risk of gastric cancer. Helicobacter. 2004;9:262–270. doi: 10.1111/j.1083-4389.2004.00221.x. [DOI] [PubMed] [Google Scholar]
- 26.Bravo LE, van Doorn LJ, Realpe JL, Correa P. Virulence-Associated Genotypes of Helicobacter pylori: Do they Explain the African Enigma? Am J Gastroenterol. 2002;97:2839–2842. doi: 10.1111/j.1572-0241.2002.07031.x. [DOI] [PubMed] [Google Scholar]
- 27.Sicinschi LA, Correa P, Peek RMJ, Camargo MC, Delgado A, Piazuelo MB, Romero-Gallo J, Bravo LE, Schneider BG. Helicobacter pylori genotyping and sequencing using paraffin-embedded biopsies from residents of Colombian areas with contrasting gastric cancer risks. Helicobacter. 2008;13:135–145. doi: 10.1111/j.1523-5378.2008.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Rugge M, Correa P, Dixon MF, Hattori T, Leandro G, Lewin K, Riddell RH, Sipponen P, Watanabe H. Gastric dysplasia - The Padova international classification. Am J Surg Pathol. 2000;24:167–176. doi: 10.1097/00000478-200002000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Taylor DE, Eaton M, Chang N, Salama SM. Construction of a Helicobacter pylori genome map and demonstration of diversity at the genome level. J Bacteriol. 1992;174:6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koehler CI, Mues MB, Dienes HP, Kriegsmann J, Schirmacher P, Odenthal M. Helicobacter pylori genotyping in gastric adenocarcinoma and MALT lymphoma by multiplex PCR analyses of paraffin wax embedded tissues. Mol Pathol. 2003;56:36–42. doi: 10.1136/mp.56.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sicinschi LA, Correa P, Peek RM, Jr, Camargo CM, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Mera R, Delgado AG, Bravo LE, Schneider BG. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2009 doi: 10.1111/j.1469-0691.2009.02811.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K, Guo Y, Ahmed SS, Toyota M, Itoh F, Suk KT, Cho MY, et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology. 2009;136:2149–2158. doi: 10.1053/j.gastro.2009.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuda H, Saito D, Hayashi S, Hisai H, Ono H, Yoshida S, Oguro Y, Noda T, Sato T, Katoh M, et al. Helicobacter pylori infection, serum pepsinogen level and gastric cancer: a case-control study in Japan. Jpn J Cancer Res. 1995;86:64–71. doi: 10.1111/j.1349-7006.1995.tb02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009 doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 36.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaise M, Yamasaki T, Yonezawa J, Miwa J, Ohta Y, Tajiri H. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacter. 2008;13:35–41. doi: 10.1111/j.1523-5378.2008.00572.x. [DOI] [PubMed] [Google Scholar]
- 38.Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, Taniguchi T. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J Biol Chem. 2000;275:22627–22630. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi T, Suzuki M, Shigematsu H, Shivapurkar N, Echebiri C, Nomura M, Stastny V, Augustus M, Wu CW, Wistuba II, Meltzer SJ, Gazdar AF. Aberrant methylation of Reprimo in human malignancies. Int J Cancer. 2005;115:503–510. doi: 10.1002/ijc.20910. [DOI] [PubMed] [Google Scholar]
- 40.Hamilton JP, Sato F, Jin Z, Greenwald BD, Ito T, Mori Y, Paun BC, Kan T, Cheng Y, Wang S, Yang J, Abraham JM, et al. Reprimo methylation is a potential biomarker of Barrett's-Associated esophageal neoplastic progression. Clin Cancer Res. 2006;12:6637–6642. doi: 10.1158/1078-0432.CCR-06-1781. [DOI] [PubMed] [Google Scholar]
- 41.Bernal C, Aguayo F, Villarroel C, Vargas M, Diaz I, Ossandon FJ, Santibanez E, Palma M, Aravena E, Barrientos C, Corvalan AH. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res. 2008;14:6264–6269. doi: 10.1158/1078-0432.CCR-07-4522. [DOI] [PubMed] [Google Scholar]
- 42.Horii A, Nakatsuru S, Ichii S, Nagase H, Nakamura Y. Multiple forms of the APC gene transcripts and their tissue-specific expression. Hum Mol Genet. 1993;2:283–287. doi: 10.1093/hmg/2.3.283. [DOI] [PubMed] [Google Scholar]
- 43.Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Laboratory Investigation. 2003;83:519–526. doi: 10.1097/01.lab.0000064704.53132.65. [DOI] [PubMed] [Google Scholar]
- 44.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, Baylin SB, Herman JG. Analysis of Adenomatous Polyposis Coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 45.Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, Motoyama T, Usuba O, Kimura W, Nishizuka S, Wilson KT, James SP, et al. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642–3646. doi: 10.1038/sj.onc.1203704. [DOI] [PubMed] [Google Scholar]
- 46.Hosoya K, Yamashita S, Ando T, Nakajima T, Itoh F, Ushijima T. Adenomatous polyposis coli 1A is likely to be methylated as a passenger in human gastric carcinogenesis. Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005;83:429–437. doi: 10.1139/o05-140. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Puisieux A, Valsesia-Wittmann S, Ansieau S. A twist for survival and cancer progression. Br J Cancer. 2006;94:13–17. doi: 10.1038/sj.bjc.6602876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamura H, Yoshida K, Haneji T. Negative regulation of TIMP1 is mediated by transcription factor TWIST1. Int J Oncol. 2009;35:181–186. doi: 10.3892/ijo_00000327. [DOI] [PubMed] [Google Scholar]
- 51.Matsuo N, Shiraha H, Fujikawa T, Takaoka N, Ueda N, Tanaka S, Nishina S, Nakanishi Y, Uemura M, Takaki A, Nakamura S, Kobayashi Y, et al. Twist expression promotes migration and invasion in hepatocellular carcinoma. BMC Cancer. 2009;9:240. doi: 10.1186/1471-2407-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasselblatt M, Mertsch S, Koos B, Riesmeier B, Stegemann H, Jeibmann A, Tomm M, Schmitz N, Wrede B, Wolff JE, Zheng W, Paulus W. TWIST-1 is overexpressed in neoplastic choroid plexus epithelial cells and promotes proliferation and invasion. Cancer Res. 2009;69:2219–2223. doi: 10.1158/0008-5472.CAN-08-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallasch C, Crabtree JE, Bevec D, Robinson PA, Wagner H, Ulrich A. Helicobacter pylori-stimulated EGF receptor transactivation requires metalloprotease cleavage of HB-EBG. Biochem Biophys Res Comm. 2002;295:695–701. doi: 10.1016/s0006-291x(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 55.Keates S, Sougioultzis S, Keates AC, D Z, Peek RM, Jr, Shaw LM, Kelly CP. cag+ Helicobacter pylori induce transactivation of the epidermal growth factor receptor in AGS gastric epithelial cells. J Biol Chem. 2001;276:48127–48134. doi: 10.1074/jbc.M107630200. [DOI] [PubMed] [Google Scholar]
- 56.Miyazaki T, Murayama Y, Shinomura Y, Yamamoto T, Watabe K, Tsutsui S, Kiyohara T, Tamura S, Hayashi N. E-cadherin gene promoter hypermethylation in H.pylori-induced enlarged fold gastritis. Helicobacter. 2007;12:523–531. doi: 10.1111/j.1523-5378.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 57.Katayama Y, Takahashi M, Kuwayama H. Helicobacter pylori causes runx3 gene methylation and its loss of expression in gastric epithelial cells, which is mediated by nitric oxide produced by macrophages. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med. 1999;190:1595–1604. doi: 10.1084/jem.190.11.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]