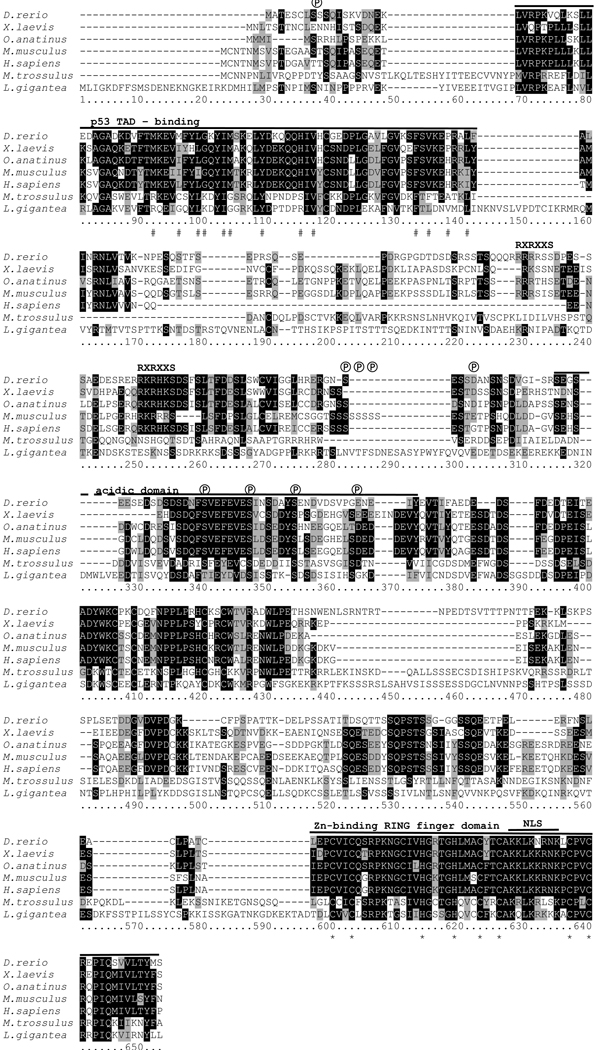
Figure 1. Multiple pairwise alignment of predicted amino acid sequences of MDM-like homologues from M. trossulus, L. gigantea (predicted from genome sequence) and of predicted MDM2 sequences from representative vertebrate species.
Shading: white on black, identical residues; black on grey, similar residues; black on white, non-homologous residues. Abbreviations: #, hydrophobic amino acids identified as making contact with p53 in the p53 binding domain of MDM2 (Kussie et al. 1996); ℗, putative conserved phosphorylation sites. RXRXXS, consensus motif for association of protein kinases, such as AKT with vertebrate MDM2, not present in molluscan MDM; NLS, nuclear localization sequence; *, conserved amino acids required for E3 activity of MDM2 within the RING-finger domain. The solid black lines indicate conserved functional domains. GenBank accession numbers are provided in Table 1.